Triple points are automatically computed during grain reconstruction. They are accessible similarly to grain boundaries as the property triplePoints of the grain list. When analyzing triple points it is a good idea to use the option removeQuadruplePoints in calcGrains to convert all quadruple points into triple points.
% import some EBSD data set
mtexdata small silent
% compute grains
grains = calcGrains(ebsd('indexed'),'removeQuadruplePoints');
%grains = calcGrains(ebsd('indexed'));
% smooth the grains a bit
grains = smooth(grains,2);
% plot the grains
plot(grains);
% extract all triple points
tP = grains.triplePoints;
% and plot the on top
hold on
plot(tP,'color','b','linewidth',2)
hold off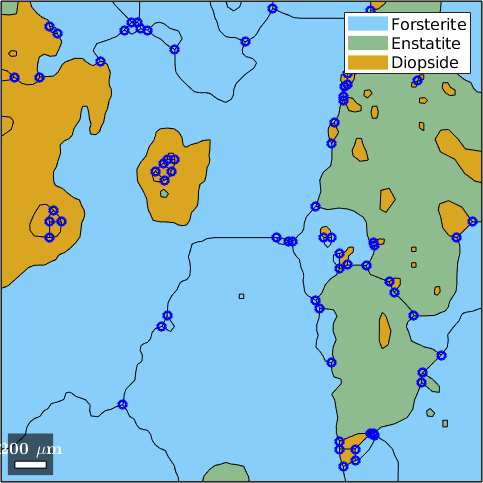
Index triple points by phase
You may index triple points by the adjacent phases. The following command gives you all triple points with at least one phase being Forsterite
tP('Forsterite')ans = triplePointList
points mineral 1 mineral 2 mineral 3
15 Forsterite Forsterite Forsterite
8 Forsterite Forsterite Enstatite
4 Forsterite Enstatite Enstatite
8 Forsterite Forsterite Diopside
19 Forsterite Enstatite Diopside
4 Forsterite Diopside DiopsideThe following command gives you all triple points with at least two phases being Forsterite
tP('Forsterite','Forsterite')ans = triplePointList
points mineral 1 mineral 2 mineral 3
15 Forsterite Forsterite Forsterite
8 Forsterite Forsterite Enstatite
8 Forsterite Forsterite DiopsideFinally, we may mark all inner Diopside triple points
hold on
plot(tP('Diopside','Diopside','Diopside'),'displayName','Di-Di-Di','color','darkred','linewidth',2)
hold off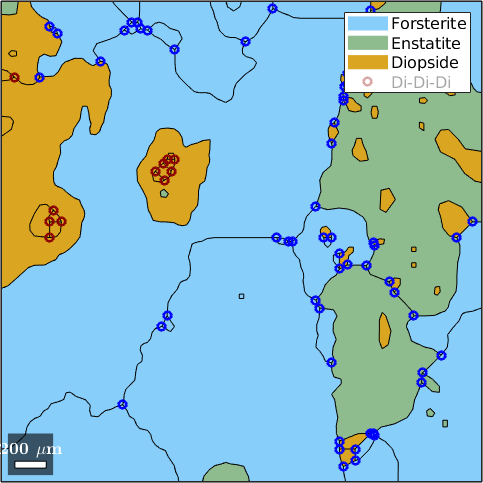
Index triple points by grains
Since, triple points are associated to grains we may single out triple points that belong to a specific grain or some subset of grains.
% find the index of the largest grain
[~,id] = max(grains.area);
% the triple points that belong to the largest grain
tP = grains(id).triplePoints;
% plot these triple points
plot(grains(id),'FaceColor',[0.2 0.8 0.8],'displayName','largest grains');
hold on
plot(grains.boundary)
plot(tP,'color','r','linewidth',2)
hold off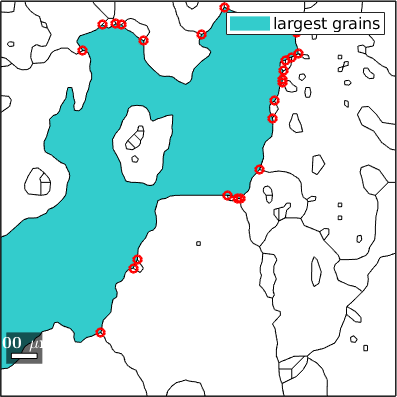
Index triple points by grain boundary
Triple points are not only a property of grains but also of grain boundaries. Thus we may ask for all triple points that belong to Fosterite - Forsterite boundaries with misorientation angle larger then 60 degree
% all Fosterite - Forsterite boundary segments
gB_Fo = grains.boundary('Forsterite','Forsterite')
% Fo - Fo segments with misorientation angle larger 60 degree
gB_large = gB_Fo(gB_Fo.misorientation.angle>60*degree)
% plot the triple points
plot(grains)
hold on
plot(gB_large,'linewidth',2,'linecolor','w')
plot(gB_large.triplePoints,'color','m','linewidth',2)
hold offgB_Fo = grainBoundary
Segments length mineral 1 mineral 2
255 10793 µm Forsterite Forsterite
gB_large = grainBoundary
Segments length mineral 1 mineral 2
55 2384 µm Forsterite Forsterite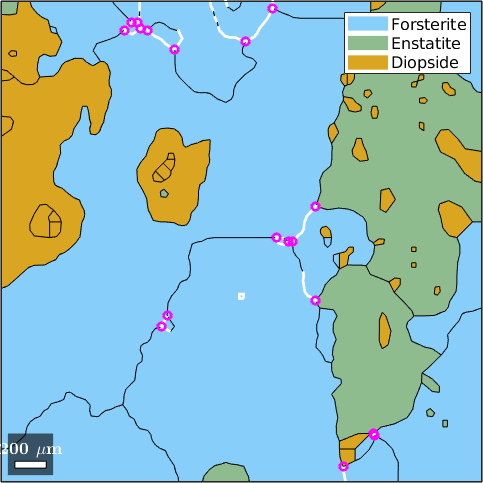
Boundary segments from triple points
On the other hand we may also ask for the boundary segments that build up a triple point. These are stored as the property boundaryId for each triple points.
% lets take Forsterite triple points
tP = grains.triplePoints('Fo','Fo','Fo');
% the boundary segments which form the triple points
gB = grains.boundary(tP.boundaryId);
% plot the triple point boundary segments
plot(grains)
hold on
plot(gB,'lineColor','w','linewidth',2)
hold off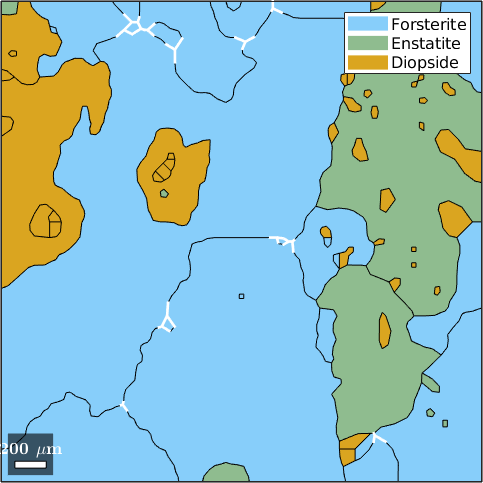
Once we have extracted the boundary segments adjacent to a triple point we may also extract the corresponding misorientations. The following command gives a n x 3 list of misorientations where n is the number of triple points
mori = gB.misorientationmori = misorientation (Forsterite → Forsterite)
size: 15 x 3
antipodal: trueHence, we can compute for each triple point the sum of misorientation angles by
sumMisAngle = sum(mori.angle,2);and my visualize it by
plot(grains,'figSize','large')
hold on
plot(tP,sumMisAngle ./ degree,'markerEdgeColor','w','MarkerSize',8)
hold off
mtexColorMap(blue2redColorMap)
setColorRange([80,180])
mtexColorbar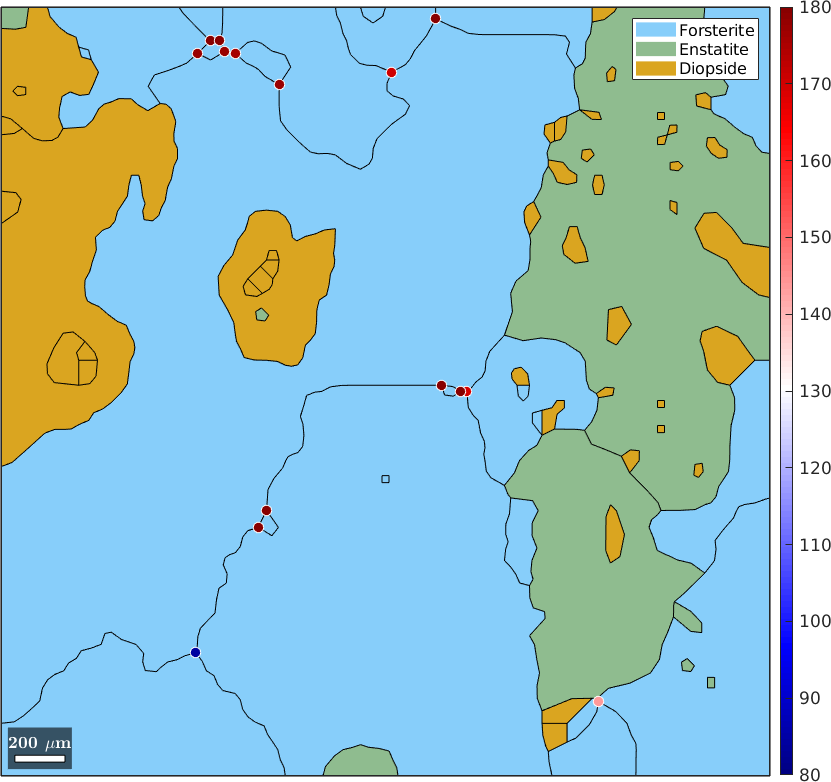
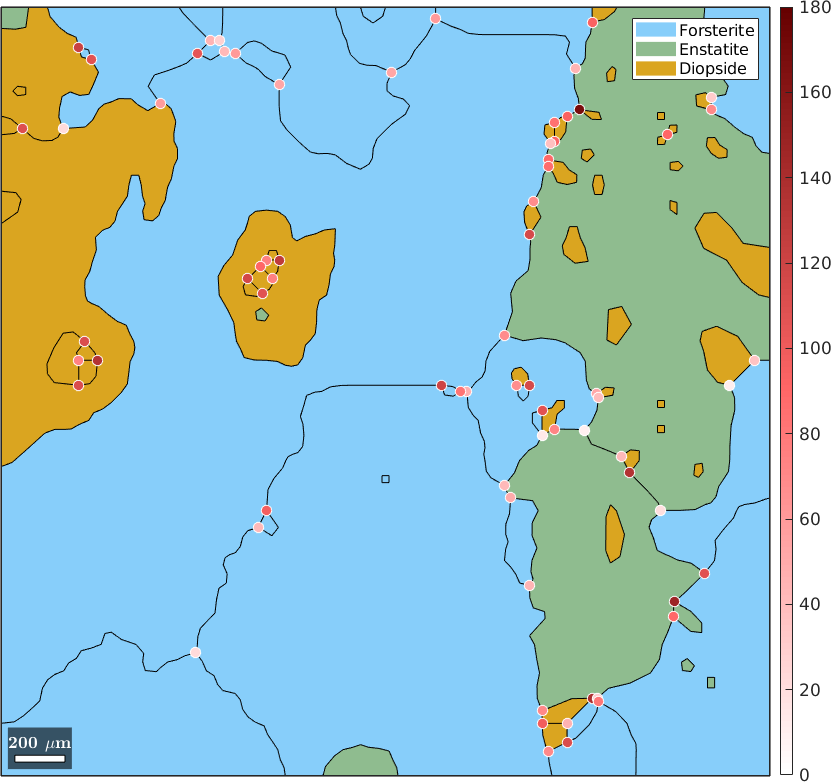
Angles at triple points
The angles at the triplepoints can be accessed by tP.angles. This is a 3 column matrix with one column for each of the three angles enclosed by the boundary segments of a triple point. Obviously, the sum of each row is always 2*pi. More interestingly is the difference between the largest and the smallest angle. Lets plot this for our test data set.
plot(grains,'figSize','large')
hold on
tP = grains.triplePoints;
plot(tP,(max(tP.angles,[],2)-min(tP.angles,[],2))./degree,'markerEdgeColor','w','MarkerSize',8)
hold off
mtexColorMap LaboTeX
setColorRange([0,180])
mtexColorbar