This section displays some of the standard orientations that are build into MTEX. The full list of predefined orientations consists of
- Cube, CubeND22, CubeND45, CubeRD
- Goss, invGoss
- Copper, Copper2
- SR, SR2, SR3, SR4
- Brass, Brass2
- PLage, PLage2, QLage, QLage2, QLage3, QLage4
For visualization we fix a generic cubic crystal symmetry and orthorhombic specimen symmetry
cs = crystalSymmetry('m-3m');
ss = specimenSymmetry('orthorhombic');and select a subset of the above predefined orientations
components = [...
orientation.goss(cs,ss),...
orientation.brass(cs,ss),...
orientation.cube(cs,ss),...
orientation.cubeND22(cs,ss),...
orientation.cubeND45(cs,ss),...
orientation.cubeRD(cs,ss),...
orientation.copper(cs,ss),...
orientation.PLage(cs,ss),...
orientation.QLage(cs,ss),...
];3d Euler angle space
Lets first visualize the orientations in the three dimensional Euler angle space
close all
for i = 1:length(components)
plot(components(i),'bunge','MarkerSize',10,'MarkerColor', ind2color(i),...
'DisplayName',round2Miller(components(i),'LaTex'))
hold on
end
legend('show','interpreter','LaTeX','location','southoutside','numColumns',3,'FontSize',1.2*getMTEXpref('FontSize'));
hold off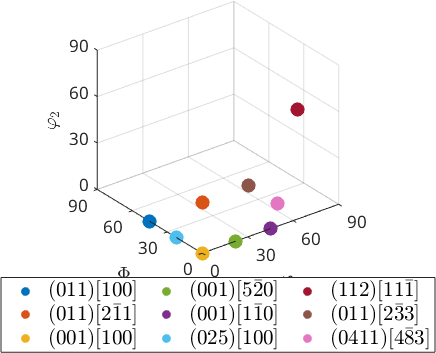
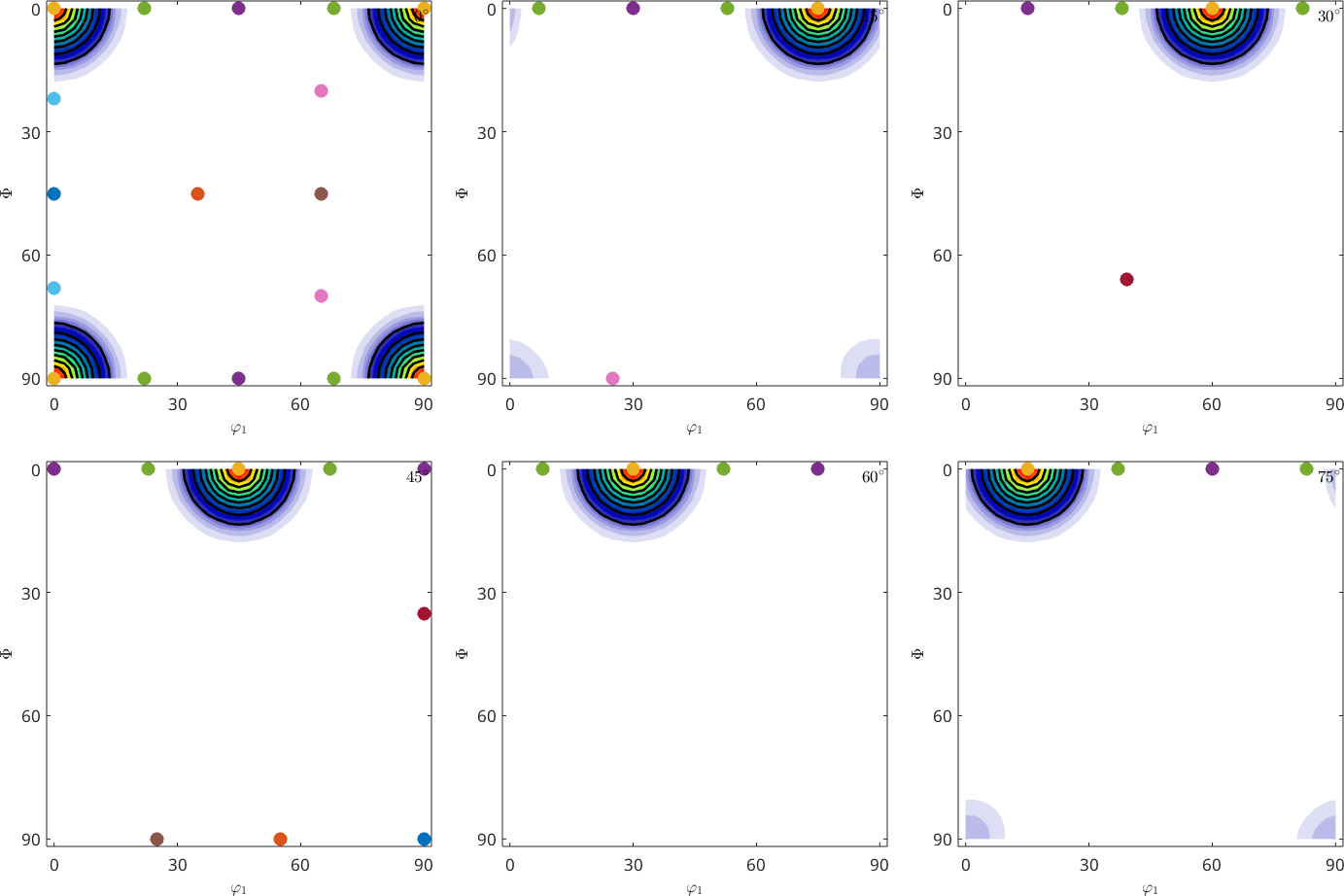
Two dimensional phi2 sections
A second common way of visualizing the orientation space are sections with fixed Euler angle phi2
close all
for i = 1:length(components)
plotSection(components(i), 'add2all', 'MarkerColor', ind2color(i),...
'DisplayName', round2Miller(components(i),'LaTex'))
end
legend('show','interpreter','LaTeX','location','southeast','FontSize',1.2*getMTEXpref('FontSize'));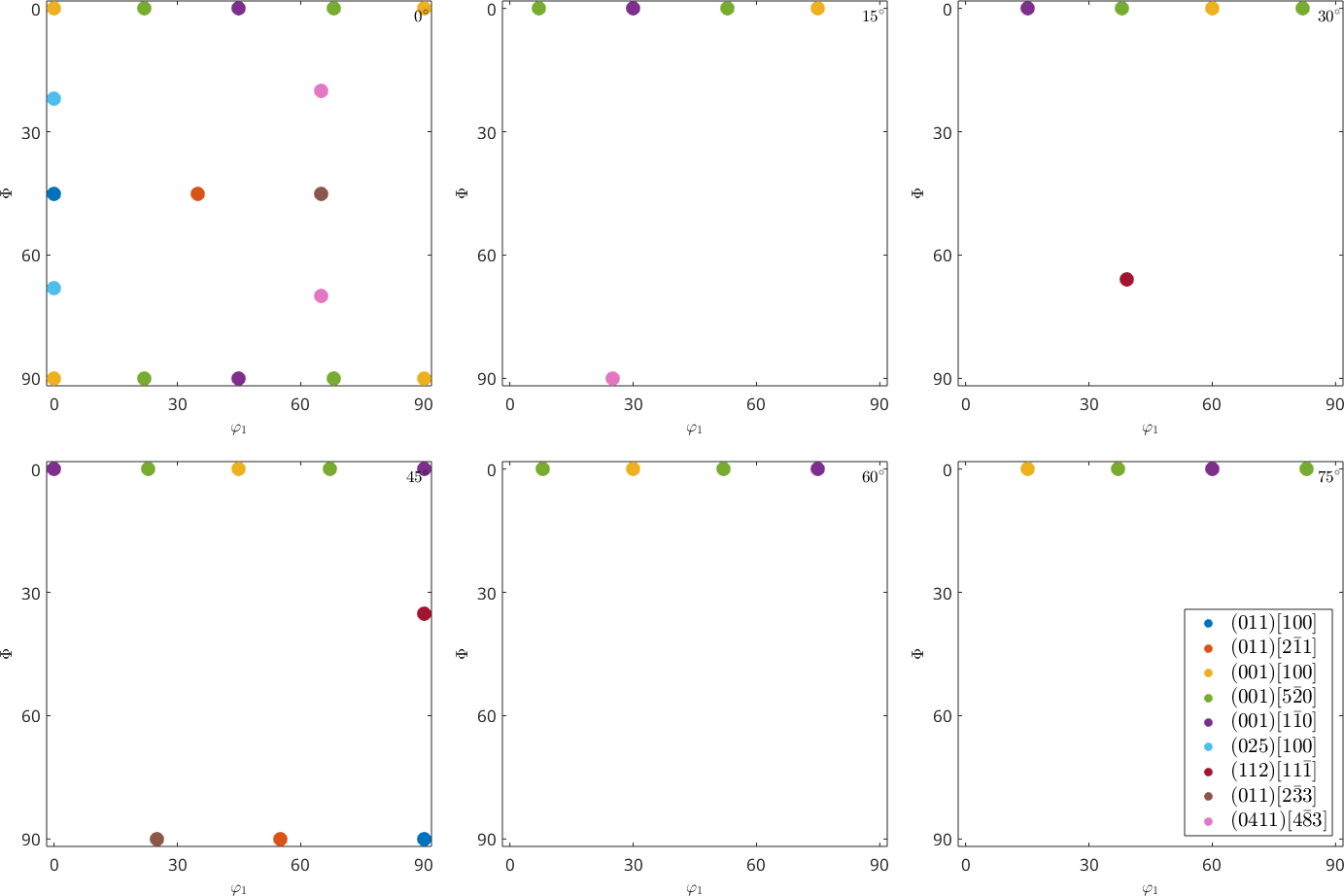
Three dimensional axis angle space
In the three dimensional axis angle space the orientation appear inside the fundamental sector
close all
for i = 1:length(components)
hold on
plot(components(i),'axisAngle','MarkerSize',10,'MarkerColor', ind2color(i),...
'DisplayName',round2Miller(components(i),'LaTex'))
axis off
end
legend('show','interpreter','LaTeX','location','southoutside','numColumns',3,'FontSize',1.2*getMTEXpref('FontSize'));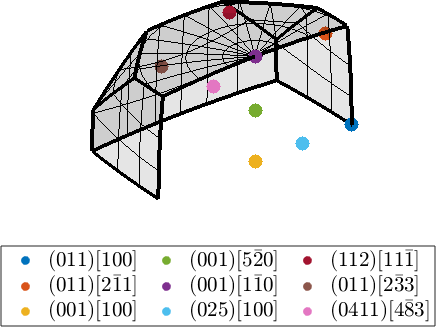
pole figures
In the major pole figures the predefined orientations appear at the following spots
plotx2north
h = Miller({1,0,0},{1,1,0},{1,1,1},{3,1,1},cs);
close all
for i = 1:length(components)
plotPDF(components(i),h,'MarkerSize',10,'MarkerColor', ind2color(i),...
'DisplayName',round2Miller(components(i),'LaTex'))
hold on
end
hold off
legend('show','interpreter','LaTeX','location','northeast','numColumns',2,'FontSize',1.2*getMTEXpref('FontSize'));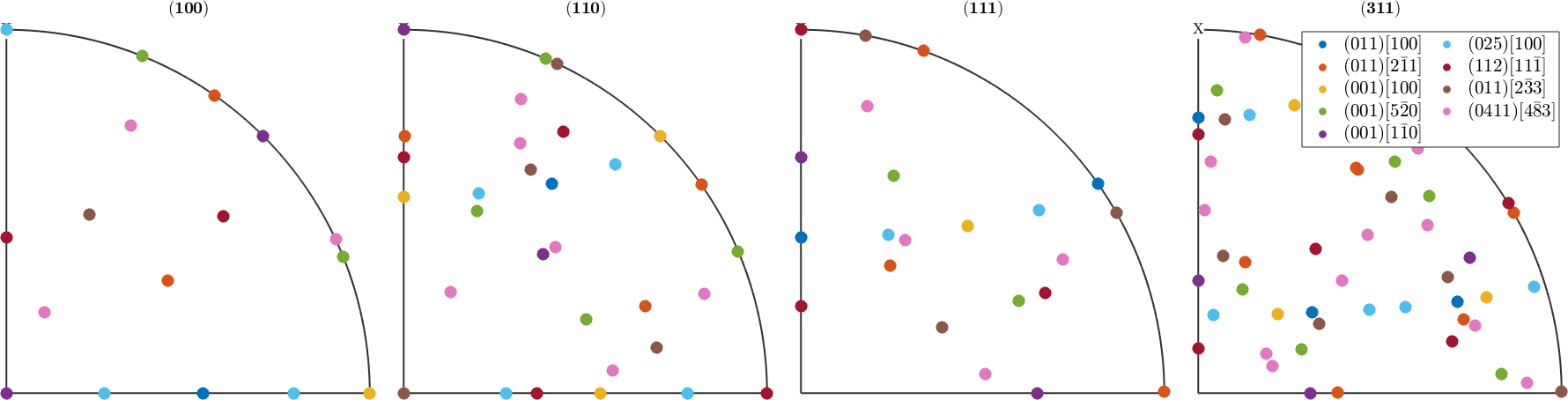
inverse pole figures
For inverse pole figure the situation is as follows. Note that the different size of the markers has been chosen to avoid overprinting.
r = [vector3d.X,vector3d.Y,vector3d.Z];
close all
for i = 1:length(components)
plotIPDF(components(i),r,'MarkerSize',(12-i)^1.5,'MarkerColor', ind2color(i),...
'DisplayName',round2Miller(components(i),'LaTex'))
hold on
end
hold off
legend('show','interpreter','LaTeX','location','northeast','numColumns',2,'FontSize',1.2*getMTEXpref('FontSize'));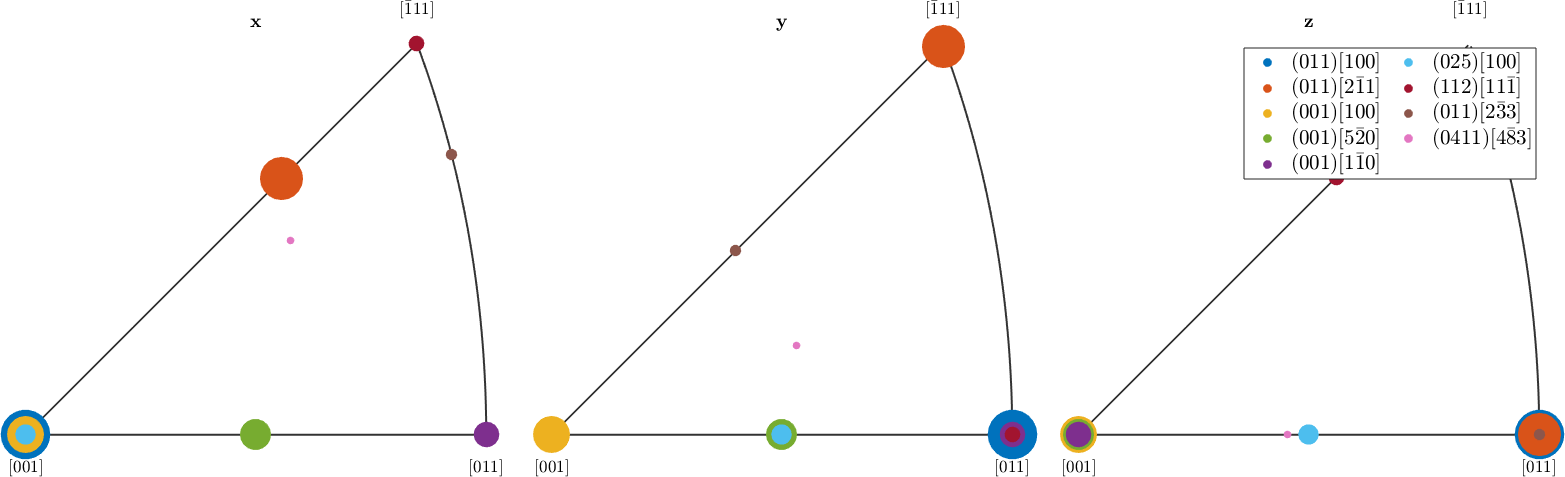
Defining an Model ODF
odf = unimodalODF(components(3),'halfwidth',7.5*degree)
plotPDF(odf,h)
hold on
plotPDF(odf,h,'contour','lineColor','k','linewidth',2)
hold offodf = SO3FunRBF (m-3m → y←↑x (mmm))
unimodal component
kernel: de la Vallee Poussin, halfwidth 7.5°
center: 1 orientations
Bunge Euler angles in degree
phi1 Phi phi2 weight
0 0 0 1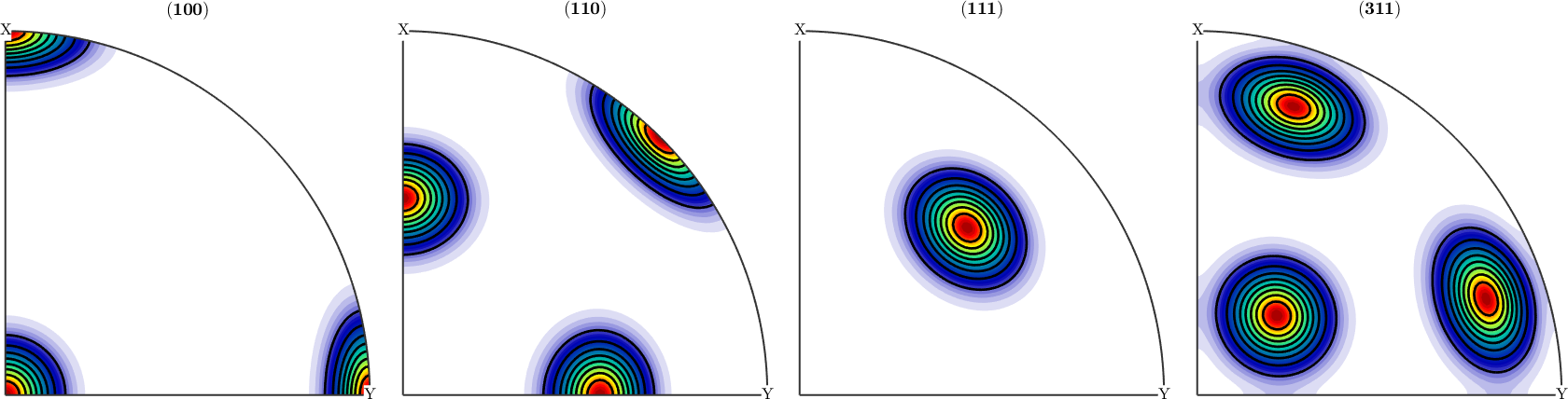
plotIPDF(odf,r)
hold on
plotIPDF(odf,r,'contour','lineColor','k','linewidth',2)
hold off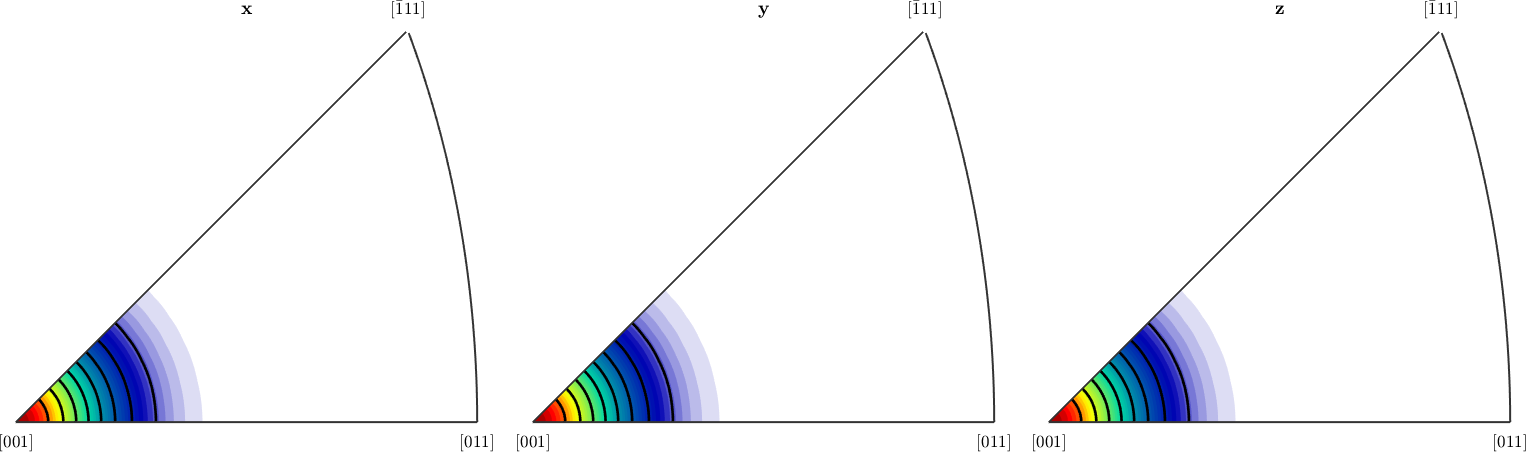
plotSection(odf)
hold on
plotSection(odf,'contour','lineColor','k','linewidth',2)
for i = 1:length(components)
plotSection(components(i),'MarkerSize',10,'filled','DisplayName',round2Miller(components(i),'LaTex'))
end
hold off