Structural conventions of the input and output of multivariate SO3FunHarmonics
In this part we deal with multivariate functions of the form
\[ f\colon \mathcal{SO}(3) \to \mathbb R^n. \]
- the structure of the nodes
@rotationsis always interpreted as a column vector - the node index is the first dimension
- the dimensions of the
SO3FunHarmonicitself is counted from the second dimension
For example we got four nodes \(R_1, R_2, R_3\) and \(R_4\) and six functions \(f_1, f_2, f_3, f_4, f_5\) and \(f_6\), which we want to store in a 3x2 array, then the following scheme applies to function evaluations:
\[ F(:, :, 1) = \pmatrix{f_1(v_1) & f_2(v_1) & f_3(v_1) \cr f_1(v_2) & f_2(v_2) & f_3(v_2) \cr f_1(v_3) & f_2(v_3) & f_3(v_3) \cr f_1(v_4) & f_2(v_4) & f_3(v_4)} \quad\mathrm{and}\quad F(:, :, 2) = \pmatrix{f_4(v_1) & f_5(v_1) & f_6(v_1) \cr f_4(v_2) & f_5(v_2) & f_6(v_2) \cr f_4(v_3) & f_5(v_3) & f_6(v_3) \cr f_4(v_4) & f_5(v_4) & f_6(v_4)}. \]
For the intern Fourier-coefficient matrix the first dimension is reserved for the Fourier-coefficients of a single function; the dimensions of the functions itself begins again with the second dimension.
If \(\bf{\hat f}_1, \bf{\hat f}_2, \bf{\hat f}_3, \bf{\hat f}_4, \bf{\hat f}_5\) and \(\bf{\hat f}_6\) would be the column vectors of the Fourier-coefficients of the functions above, internally they would be stored in \(\hat F\) as follows. \[ \hat F(:, :, 1) = \pmatrix{\bf{\hat f}_1 & \bf{\hat f}_2 & \bf{\hat f}_3} \quad\mathrm{and}\quad \hat F(:, :, 2) = \pmatrix{\bf{\hat f}_4 & \bf{\hat f}_5 & \bf{\hat f}_6}. \]
Defining a multivariate SO3FunHarmonic
Definition via function values
At first we need some vertices
nodes = equispacedSO3Grid(crystalSymmetry,specimenSymmetry,'points',1e5);
nodes = nodes(:);Next we define function values for the vertices
y = [SO3Fun.dubna(nodes), (nodes.a .* nodes.b).^(1/4)];
nodes.CS = SO3Fun.dubna.CS;Now the actual command to get a (2x1) SO3F1 of type \(~\) SO3FunHarmonic is
SO3F1 = SO3FunHarmonic.interpolate(nodes, y,'maxit',10)Warning: Maximum number of iterations reached, result may not
have converged to the optimum yet.
SO3F1 = SO3FunHarmonic (Quartz → y↑→x)
isReal: false
size: 2 x 1
bandwidth: 54
weights: [0.96 0.47]It is also possible to interpolate one component by an SO3FunRBF, that means
SO3F2 = interp(nodes,y(:,1))Warning: Maximum number of iterations reached, result may not
have converged to the optimum yet.
SO3F2 = SO3FunRBF (Quartz → y↑→x)
multimodal components
kernel: de la Vallee Poussin, halfwidth 5°
center: 19848 orientations, resolution: 5°
weight: 1This is only possible for univariate functions.
Definition via function handle
If we have a function handle for the function we could create a SO3FunHarmonic via quadrature. At first let us define a function handle which takes \(~\) rotation as an argument and returns double:
f = @(rot) [exp(rot.a+rot.b+rot.c)+50*(rot.b-cos(pi/3)).^3.*(rot.b-cos(pi/3) > 0), rot.a, rot.b, rot.c];Now we call the quadrature command to get (4x1) SO3F3 of type SO3FunHarmonic
SO3F3 = SO3FunHarmonic.quadrature(f, 'bandwidth', 50,SO3F1.CS)SO3F3 = SO3FunHarmonic (Quartz → y↑→x)
isReal: false
size: 4 x 1
bandwidth: 50Definition via Fourier-coefficients
If we already know the Fourier-coefficients, we can simply hand them in the format above to the constructor of SO3FunHarmonic.
SO3F4 = SO3FunHarmonic(eye(10))SO3F4 = SO3FunHarmonic (y↑→x → y↑→x)
isReal: false
size: 10 x 1
bandwidth: 1- This command stores the ten first Wigner-D functions in
SO3F4
Operations which differ from an univariate SO3FunHarmonic
Some default matrix and vector operations
You can concatenate and refer to functions as MATLAB does with vectors and matrices
SO3F5 = [SO3F1; SO3F3];
SO3F5(2:4)ans = SO3FunHarmonic (Quartz → y↑→x)
isReal: false
size: 3 x 1
bandwidth: 54
weights: [0.47 1.1 -0.25]You can conjugate the Fourier-coefficients and transpose/ctranspose the multivariate SO3FunHarmonic.
conj(SO3F1);
SO3F1.';
SO3F1'ans = SO3FunHarmonic (Quartz → y↑→x)
isReal: false
size: 1 x 2
bandwidth: 54
weights: [0.96 0.47]Some other operations
length(SO3F1)
size(SO3F3)
SO3F4 = reshape(SO3F4, 2, [])ans =
2
ans =
4 1
SO3F4 = SO3FunHarmonic (y↑→x → y↑→x)
isReal: false
size: 2 x 5
bandwidth: 1
sum and mean
If we do not specify further options to sum or mean they give we the integral or the mean value back for each function. You could also calculate the conventional sum or the meanvalue over a dimension of a multivariate SO3FunHarmonic.
sum(SO3F1, 1)
mean(SO3F4, 2)ans = SO3FunHarmonic (Quartz → y↑→x)
isReal: false
bandwidth: 54
weight: 1.43+0.2i
ans = SO3FunHarmonic (y↑→x → y↑→x)
antipodal: true
size: 2 x 1
bandwidth: 1
weights: [0.2 0]min/max
If the min or max command gets a multivariate SO3FunHarmonic the pointwise minimum or maximum is calculated along the first non-singelton dimension if not specified otherwise.
Therefore the function has to be real valued
SO3F4.isReal = 1;
min(SO3F4)ans =
Columns 1 through 7
1.0000 -1.7320 -1.2247 -1.7320 -1.2247 -1.7321 -1.2247
Columns 8 through 10
-1.7320 -1.2247 -1.7320Remark on the matrix product
At this point the matrix product is implemented pointwise and not as the usual matrix product.
SO3F1.CS=specimenSymmetry;
SO3F1 .* SO3F4ans = SO3FunHarmonic (y↑→x → y↑→x)
isReal: false
size: 2 x 5
bandwidth: 55Visualization of multivariate SO3FunHarmonic
Similarly to the univariate case we also can look at the Fourier coefficients of multivariate functions.
plotSpektra(SO3F3)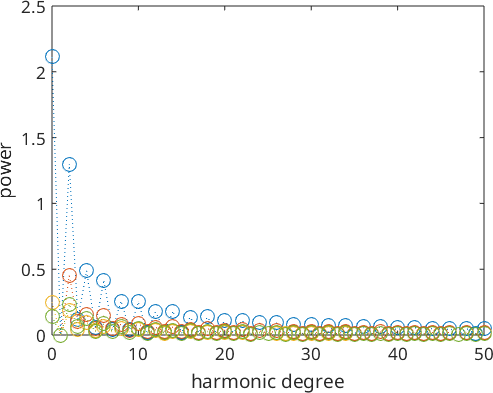
The section plot and the 3d plot are performed only for the first component of a multivariate function
plot(SO3F3)Warning: You try to plot an multivariate function. Plot the
desired components manually. In the following the first component
is plotted.
Warning: Imaginary part of complex valued SO3FunHarmonic is
ignored. In the following only the real part is plotted.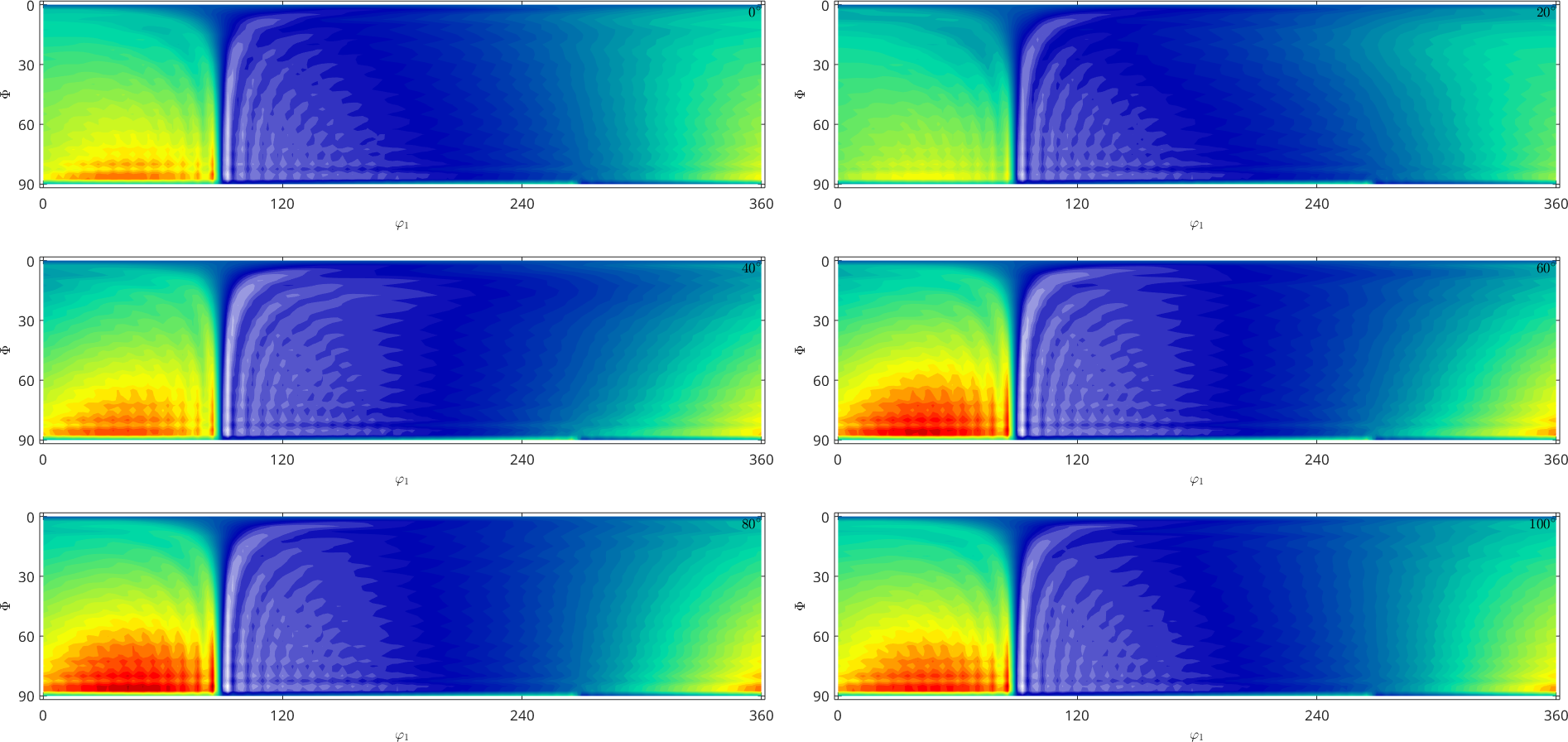
plot3d(SO3F3)Warning: You try to plot an multivariate function. Plot the
desired components manually. In the following the first component
is plotted.
Warning: Imaginary part of complex valued SO3FunHarmonic is
ignored. In the following only the real part is plotted.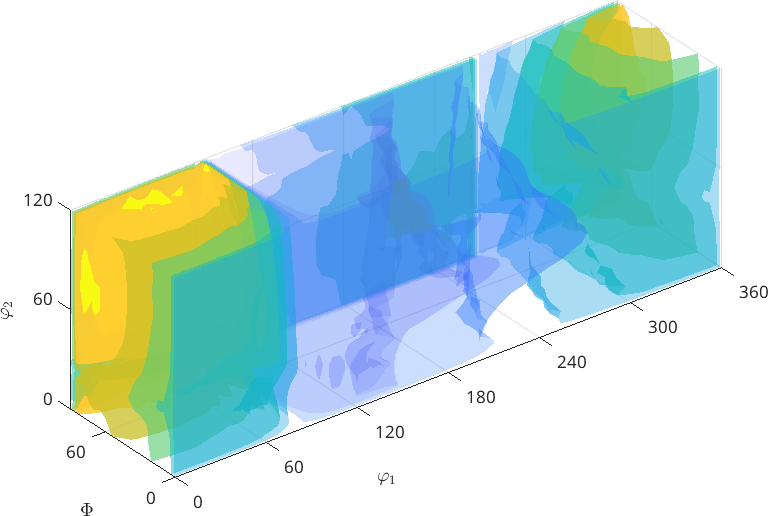
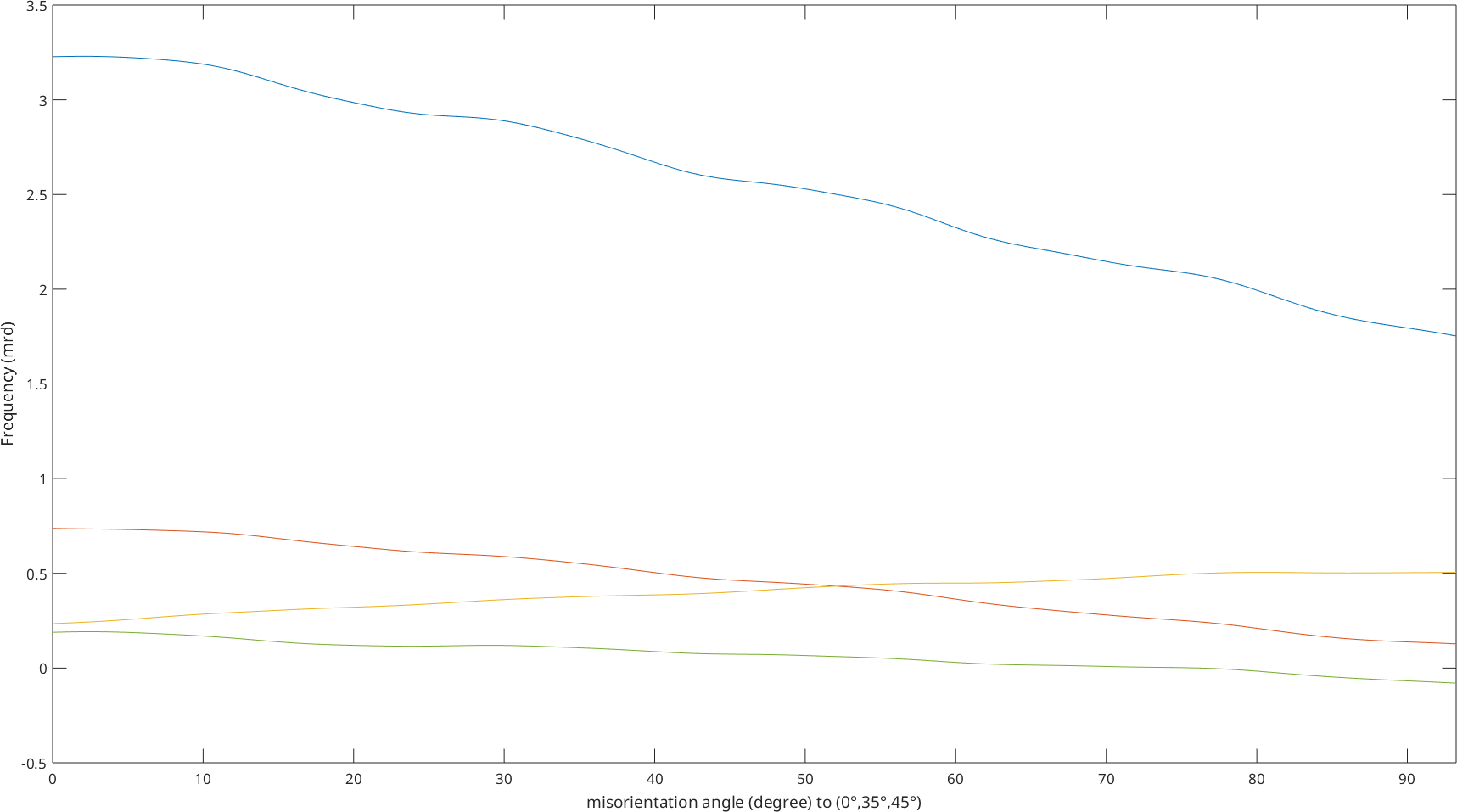
while the plot along a specific fibre includes all components.
plotFibre(SO3F3,fibre.beta)Warning: Imaginary part of complex valued SO3FunHarmonic is
ignored. In the following only the real part is plotted.