In this section we discuss how to manipulate pole figure data, e.g., to correct for measurement errors. To illustrate lets start by importing some sample XRD data.
mtexdata geesthacht
% plot imported pole figure
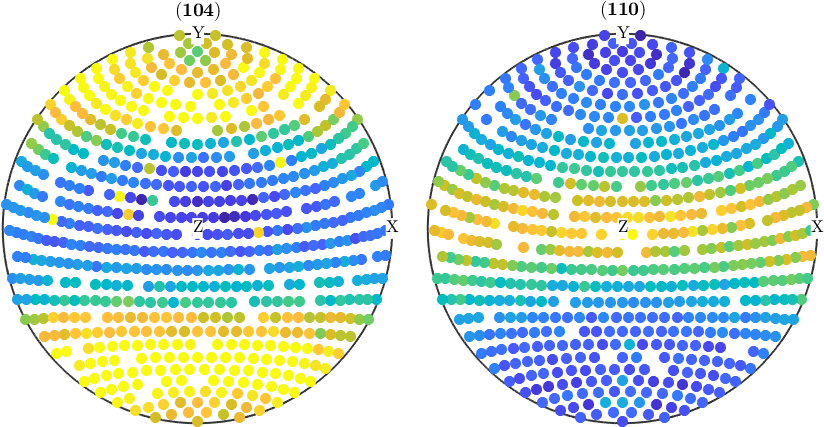
plot(pf)pf = PoleFigure (y↑→x)
crystal symmetry : m-3m
h = (104), r = 679 x 1 points
h = (104), r = 16 x 1 points
h = (110), r = 679 x 1 points
h = (110), r = 16 x 1 points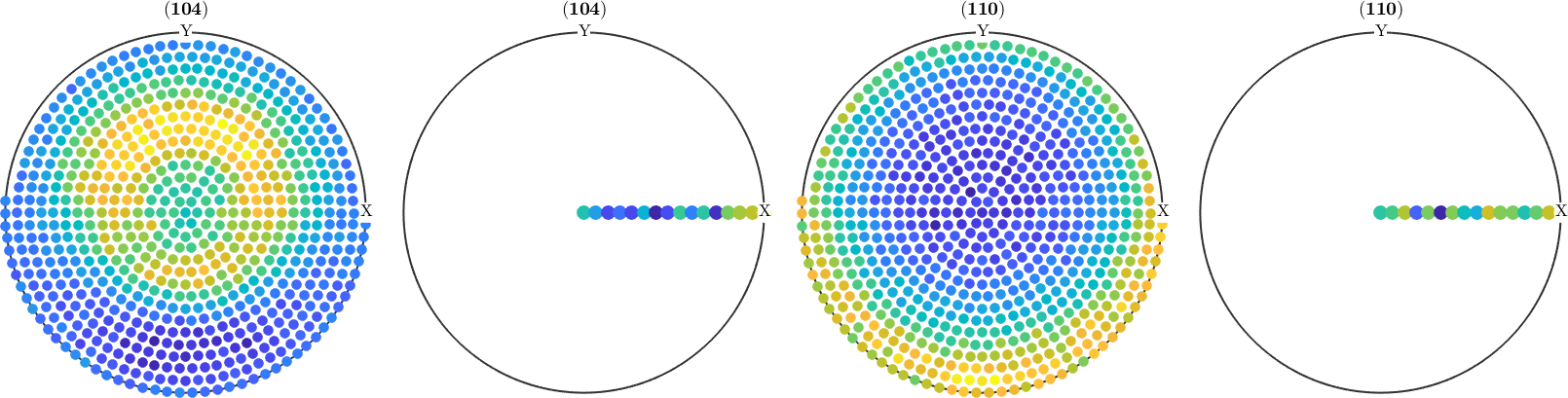
Splitting and Reordering of Pole Figures
As we can see the first and the third pole figure complete pole figures and the second and the fourth pole figures contain some values for background correction. Let us, therefore, split the pole figures into these two groups.
pf_complete = pf({1,3})
pf_background= pf({2,4})pf_complete = PoleFigure (y↑→x)
crystal symmetry : m-3m
h = (104), r = 679 x 1 points
h = (110), r = 679 x 1 points
pf_background = PoleFigure (y↑→x)
crystal symmetry : m-3m
h = (104), r = 16 x 1 points
h = (110), r = 16 x 1 pointsActually, it is possible to work with pole figures as with simple numbers. E.g. it is possible to add / subtract pole figures. A superposition of the first and the third pole figures can be written as
2*pf({1}) + 3*pf({3})ans = PoleFigure (y↑→x)
crystal symmetry : m-3m
h = (104)(110), r = 679 x 1 pointsCorrect pole figure data
In order to correct pole figures for background radiation and defocusing one can use the command correct. In our case the syntax is
pf = correct(pf_complete,'background',pf_background);
plot(pf)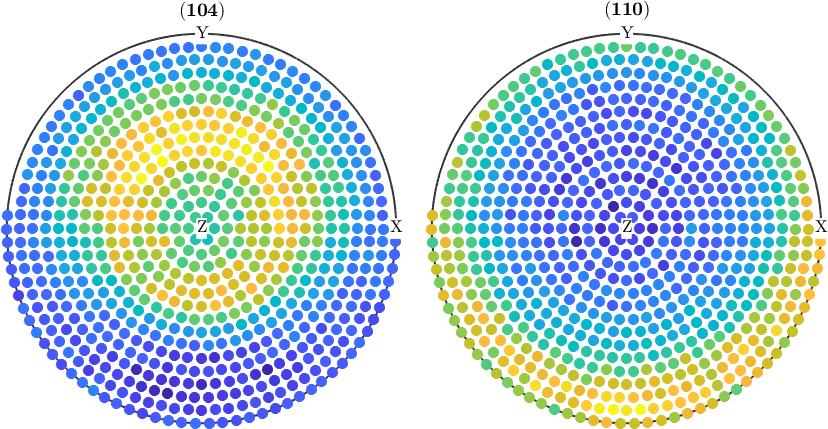
Normalize pole figures
Sometimes people want to have normalized pole figures. In the case of complete pole figures, this can be simply archived using the command normalize
pf_normalized = normalize(pf);
plot(pf_normalized)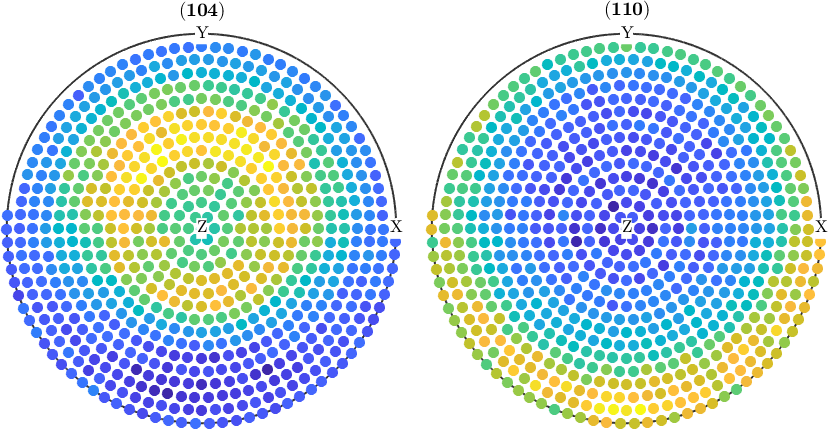
However, in the case of incomplete pole figures, it is well known, that the normalization can only by computed from an ODF. Therefore, one has to proceed as follows:
% compute an ODF from the pole figure data
odf = calcODF(pf);
% and use it for normalization
pf_normalized = normalize(pf,odf);
plot(pf_normalized)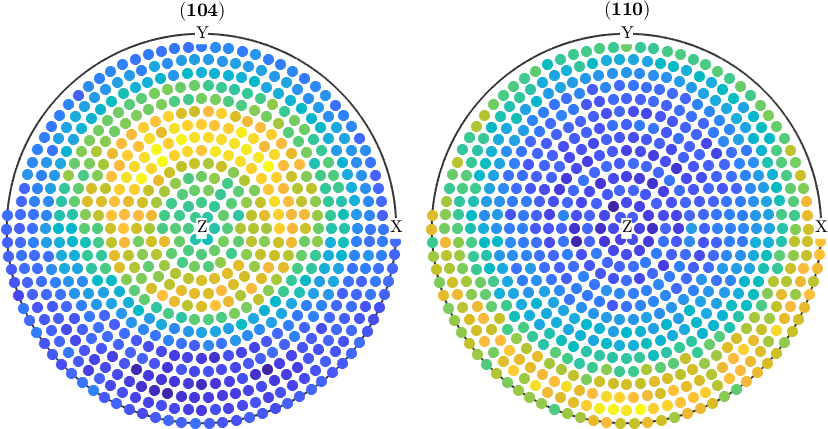
Modify certain pole figure values
As pole figures are usually experimental data they may contain outliers. In order to remove outliers from pole figure data one can use the function isOutlier. Here a simple example:
% Let us add 100 random outliers to the pole figure data
% First we select 100 random positions within the pole figures
ind = randperm(pf.length,100);
% Next we multiply the intensity at these positions by a random value
% between 3 and 4
factor = 3+rand(100,1);
pf(ind).intensities = pf(ind).intensities(:) .* factor;
% Let's check the result
plot(pf)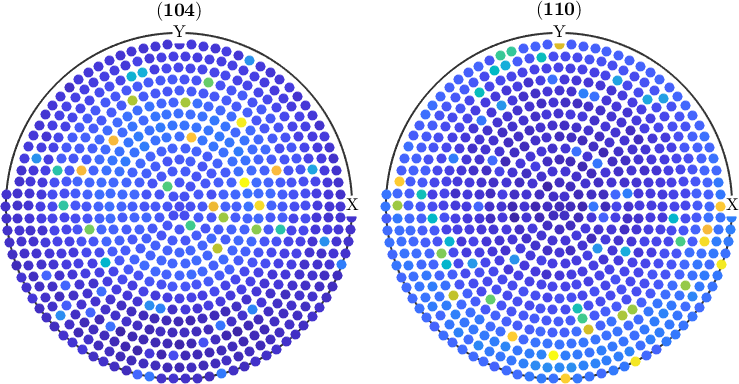
check for outliers
condition = pf.isOutlier;
% remove outliers
pf(condition) = [];
% plot the corrected pole figures
plot(pf)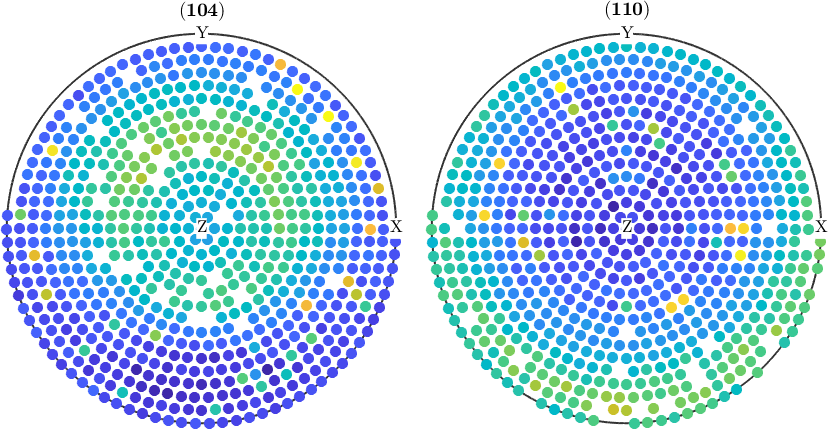
Sometimes applying the above correction is not sufficient. Then it can help to repeat the outlier detection ones again
pf(pf.isOutlier) = [];
plot(pf)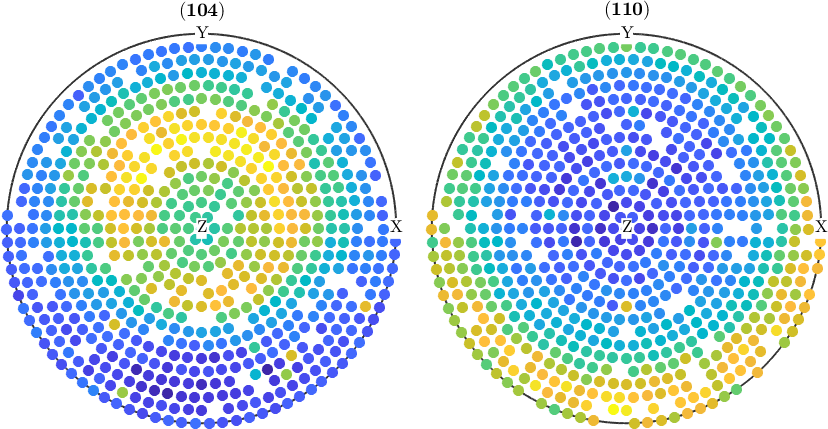
Remove certain measurements from the data
In the same way, as we removed the outlier one can manipulate and delete pole figure data by any criteria. Lets, e.g. cap all values that are larger than 500.
% find those values
condition = pf.intensities > 500;
% cap the values in the pole figures
pf(condition).intensities = 500;
plot(pf)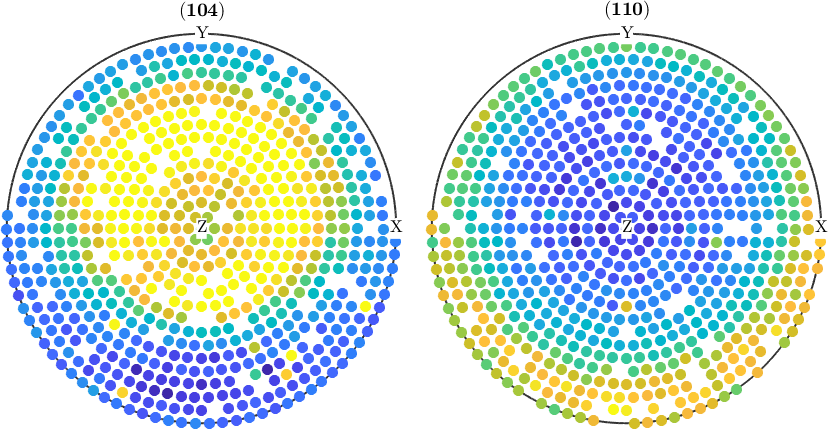
Rotate pole figures
Sometimes it is necessary to rotate the pole figures. In order to do this with MTEX one has first to define a rotation, e.e. by
% This defines a rotation around the x-axis about 100 degree
rot = rotation.byAxisAngle(xvector,100*degree);Second, the command rotate can be used to rotate the pole figure data.
pf_rotated = rotate(pf,rot);
plot(pf_rotated,'antipodal')