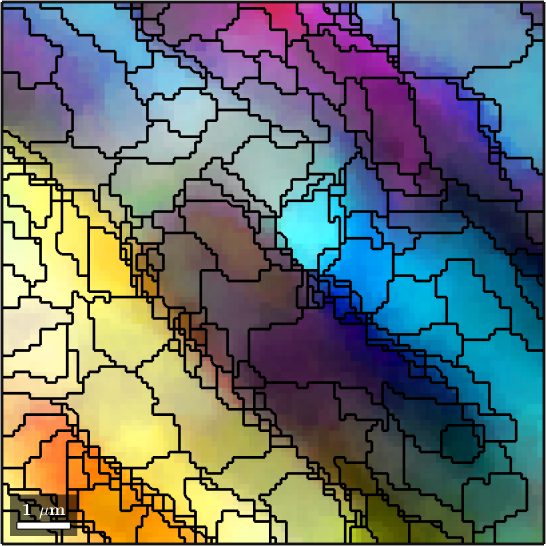
1) no fill, no grains, all pixels
mtexdata small
[grains,ebsd.grainId,ebsd.mis2mean] = calcGrains(ebsd);
ebsd(grains(grains.numPixel<=3)) = [];
[grains,ebsd.grainId,ebsd.mis2mean] = calcGrains(ebsd);
F = splineFilter;
ebsd = smooth(ebsd,F);
[grains,ebsd.grainId,ebsd.mis2mean] = calcGrains(ebsd);
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold offebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)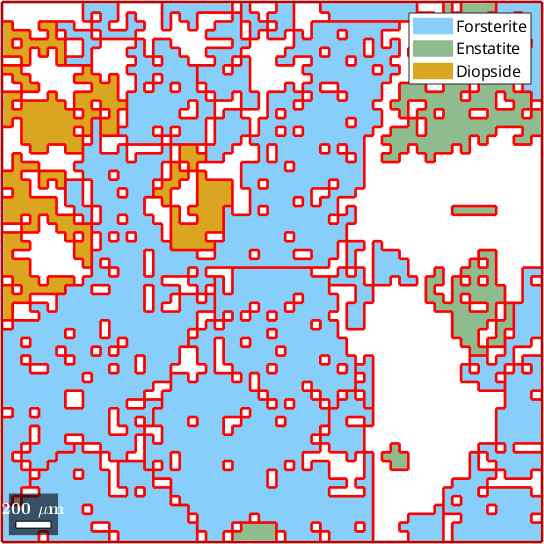
2) no fill, no grains, indexed pixels
mtexdata small
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
ebsd(grains(grains.numPixel<=3)) = [];
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
F = splineFilter;
ebsd = smooth(ebsd('indexed'),F);
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
nextAxis
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold offebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)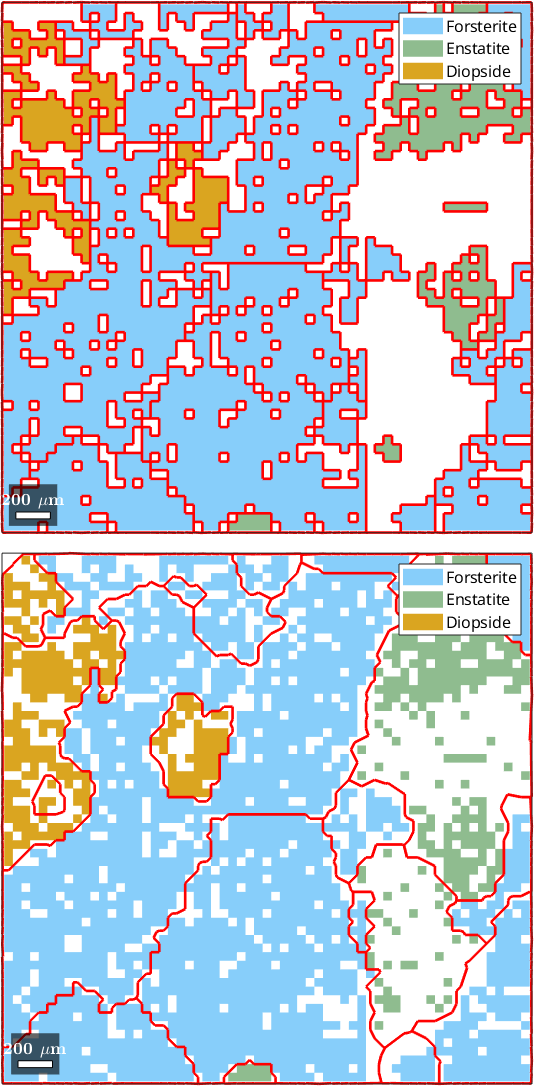
3) fill, no grains, all pixels
mtexdata small
[grains,ebsd.grainId] = calcGrains(ebsd);
ebsd(grains(grains.numPixel<=3)) = [];
[grains,ebsd.grainId] = calcGrains(ebsd);
F = splineFilter;
ebsd = smooth(ebsd,F,'fill');
[grains,ebsd.grainId] = calcGrains(ebsd);
nextAxis
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold offebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)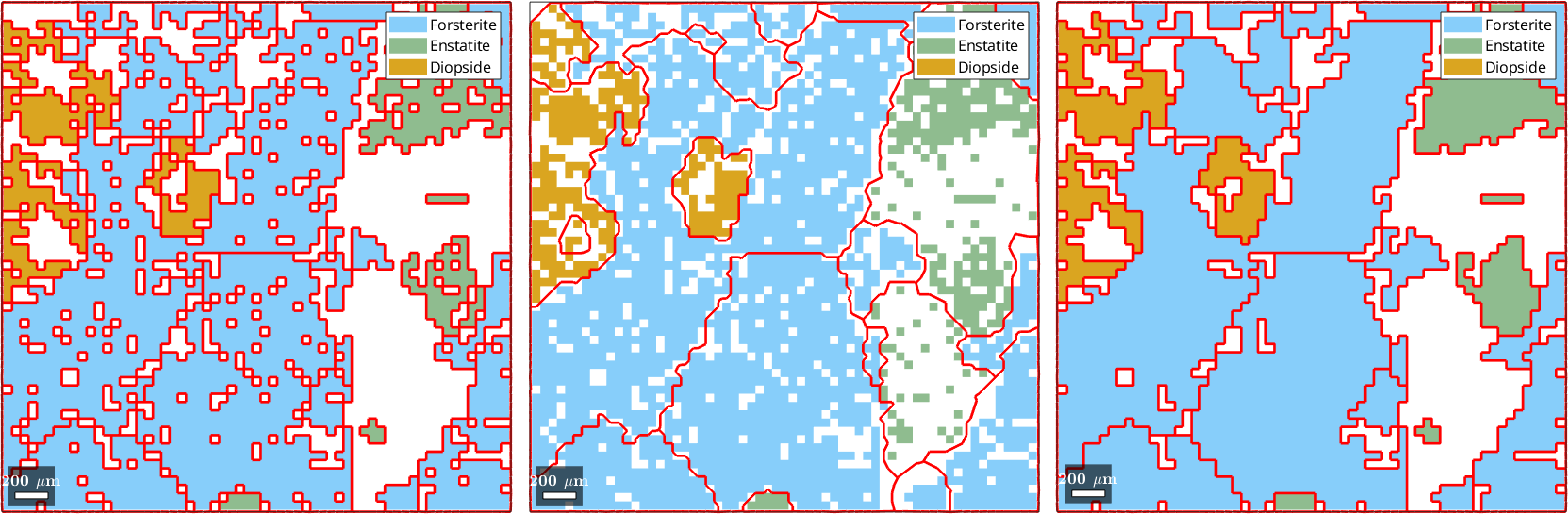
4) fill, no grains, indexed pixels
mtexdata small
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
ebsd(grains(grains.numPixel<=3)) = [];
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
F = splineFilter;
ebsd = smooth(ebsd('indexed'),F,'fill');
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
nextAxis
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold offebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)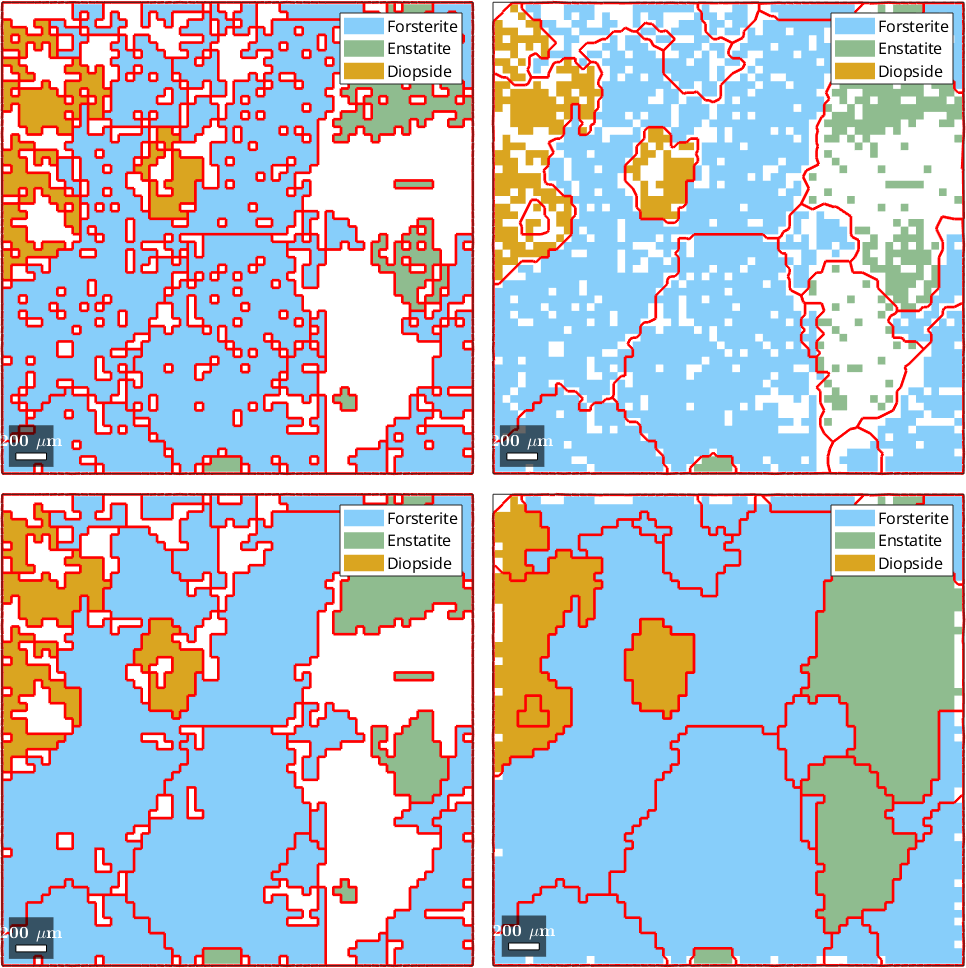
5) fill, grains, indexed pixels
mtexdata small
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
ebsd(grains(grains.numPixel<=3)) = [];
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
F = splineFilter;
ebsd = smooth(ebsd('indexed'),F,'fill',grains);
[grains,ebsd('indexed').grainId] = calcGrains(ebsd('indexed'));
nextAxis
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold off
drawNow(gcm)ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)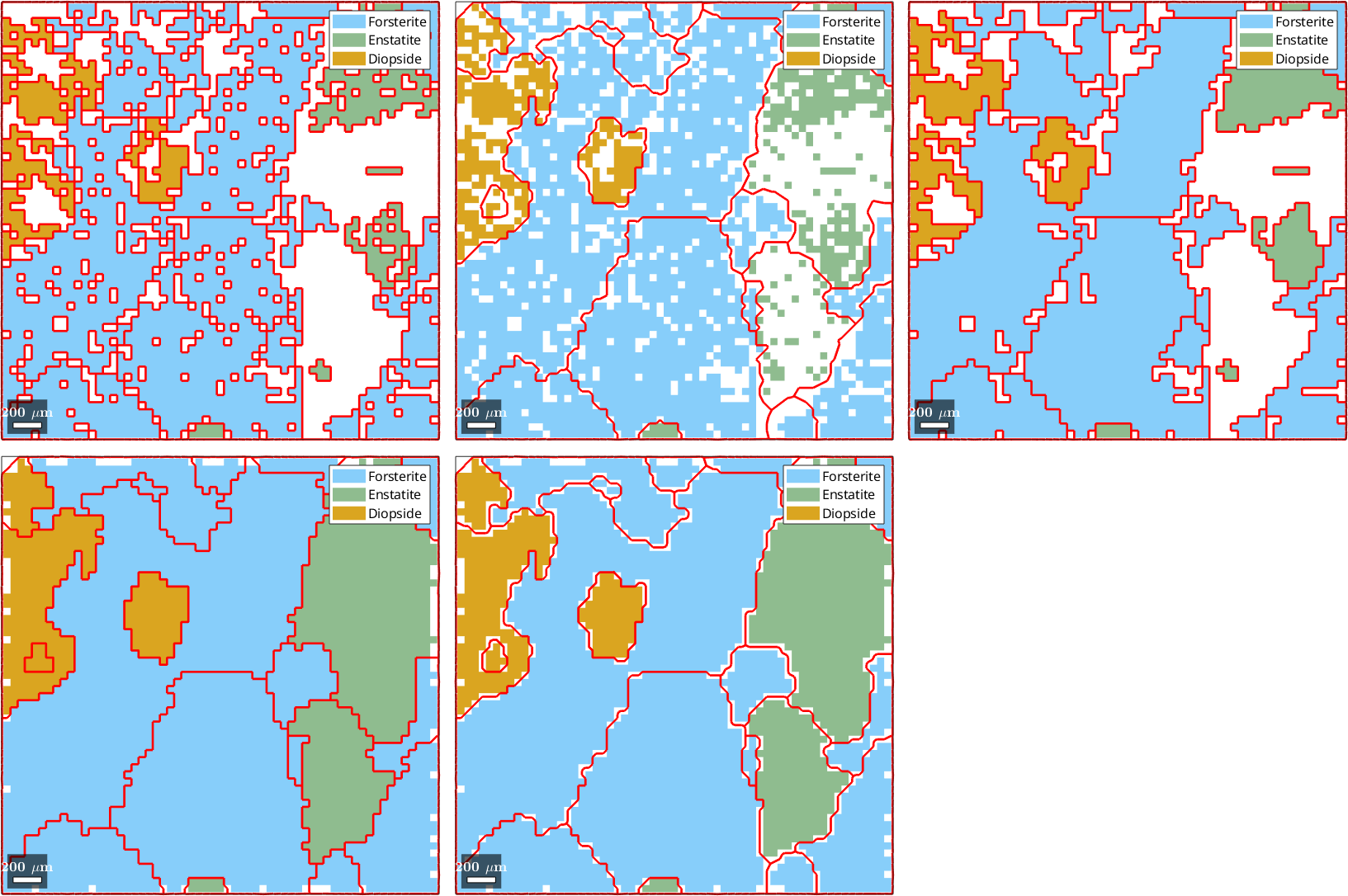
6) Multiscale clustering method
mtexdata small
grains = calcGrains(ebsd('indexed'),'FMC',1)
nextAxis
plot(ebsd)
hold on
plot(grains.boundary,'lineColor',[1 0 0],'linewidth',2)
hold off
drawNow(gcm)ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 1197 (32%) notIndexed
1 1952 (52%) Forsterite LightSkyBlue mmm
2 290 (7.8%) Enstatite DarkSeaGreen mmm
3 282 (7.6%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [33000, 36000] x [4500, 7500] x [0, 0]
Normal vector: (0,0,1)
grains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 23 1952 Forsterite mmm
2 7 290 Enstatite mmm
3 38 282 Diopside 12/m1 X||a*, Y||b*, Z||c
boundary segments: 1177 (53903 µm)
inner boundary segments: 0 (0 µm)
triple points: 60
Properties: meanRotation, GOS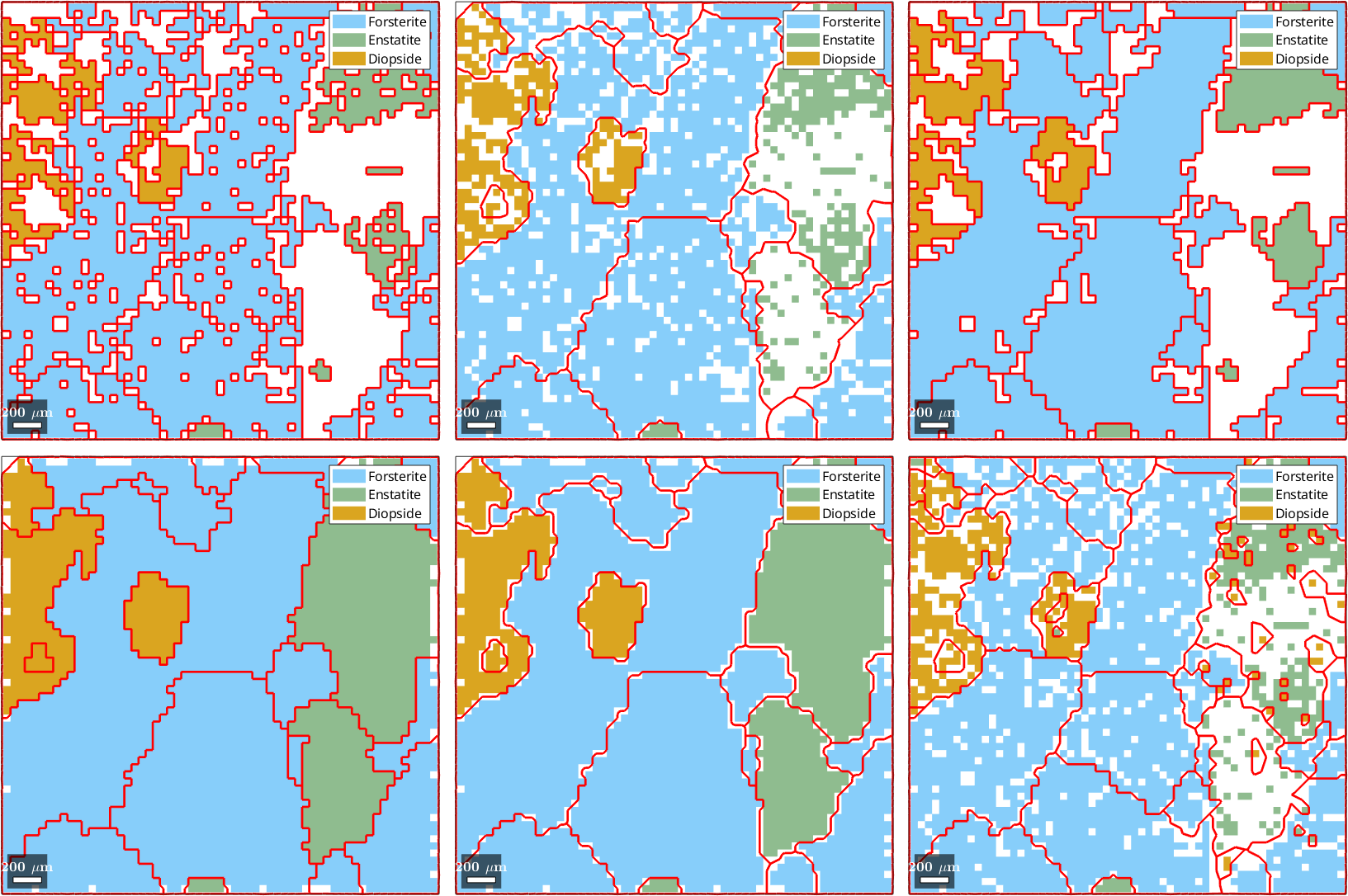
Multiscale clustering method
When analyzing grains with gradual and subtle boundaries the threshold based method may not lead to the desired result.
Let us consider the following example
mtexdata single
colorKey = axisAngleColorKey(ebsd);
colorKey.oriRef = mean(ebsd.orientations);
plot(ebsd,colorKey.orientation2color(ebsd.orientations))ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
1 10201 (100%) Al LightSkyBlue m-3m
Scan unit : um
X x Y x Z : [6, 16] x [4, 14] x [0, 0]
Normal vector: (0,0,1)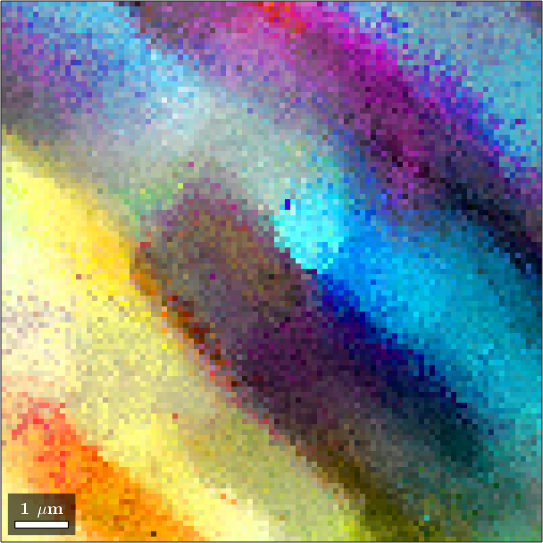
We obeserve that the are no rapid changes in the orientation which would allow for applying the threshold based algorithm. Setting the threshold angle to a very small value would include many irrelevant or false regions.
grains_high = calcGrains(ebsd,'angle',1*degree);
grains_low = calcGrains(ebsd,'angle',0.5*degree);
figure
plot(ebsd,colorKey.orientation2color(ebsd.orientations))
hold on
plot(grains_high.boundary)
hold off
figure
plot(ebsd,colorKey.orientation2color(ebsd.orientations))
hold on
plot(grains_low.boundary)
hold off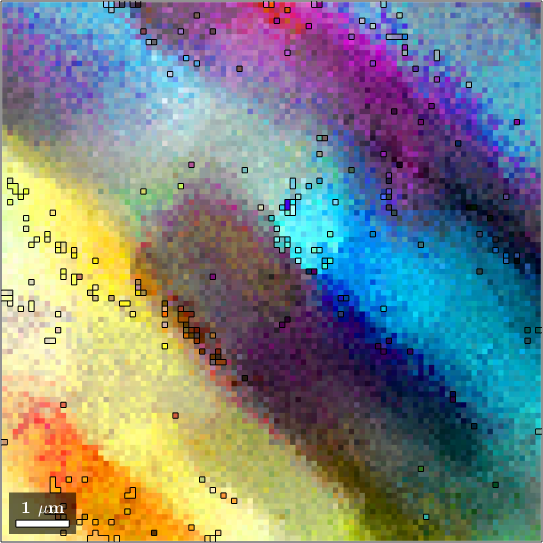
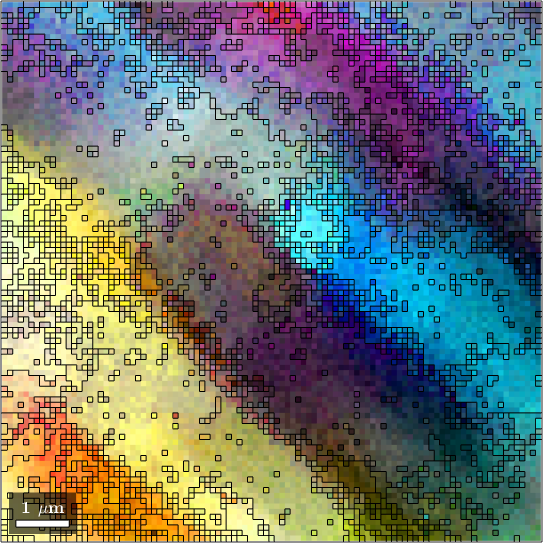
As an alternative MTEX includes the fast multi-scale clustering method (FMC method) which constructs clusters in a hierarchical manner from single pixels using fuzzy logic to account for local, as well as global information.
Analogous with the threshold angle, a single parameter, C_Maha controls the sensitivity of the segmentation. A C_Maha value of 3.5 properly identifies the sub-grain features. A C_Maha value of 3 captures more general features, while a value of 4 identifies finer features but is slightly oversegmented.
grains_FMC = calcGrains(ebsd('indexed'),'FMC',3.8)
grains = calcGrains(ebsd('indexed'))
% smooth grains to remove staircase effect
grains_FMC = smooth(grains_FMC);grains_FMC = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 17 10201 Al m-3m
boundary segments: 1937 (156 µm)
inner boundary segments: 0 (0 µm)
triple points: 14
Id Phase Pixels meanRotation GOS
1 1 7 (317°,128°,159°) 0.0084
2 1 1120 (239°,74°,319°) 0.025
3 1 517 (238°,75°,320°) 0.01
4 1 1783 (56°,103°,40°) 0.022
5 1 11 (57°,104°,220°) 0.0074
6 1 2 (237°,79°,322°) 0.0028
7 1 7 (343°,43°,249°) 0.008
8 1 989 (236°,79°,322°) 0.013
9 1 1060 (239°,75°,320°) 0.02
10 1 76 (238°,75°,320°) 0.0079
11 1 1556 (238°,77°,320°) 0.022
12 1 349 (237°,77°,320°) 0.011
13 1 401 (342°,41°,251°) 0.01
14 1 402 (238°,77°,320°) 0.012
15 1 184 (237°,77°,319°) 0.013
16 1 957 (316°,129°,74°) 0.018
17 1 780 (238°,78°,321°) 0.012
grains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 1 10201 Al m-3m
boundary segments: 789 (41 µm)
inner boundary segments: 0 (0 µm)
triple points: 0
Id Phase Pixels meanRotation GOS
1 1 10201 (237°,77°,320°) 0.0344809We observe how this method nicely splits the measurements into clusters of similar orientation
%plot(ebsd,oM.orientation2color(ebsd.orientations))
plot(ebsd,colorKey.orientation2color(ebsd.orientations))
% start override mode
hold on
plot(grains_FMC.boundary,'linewidth',1.5)
% stop override mode
hold off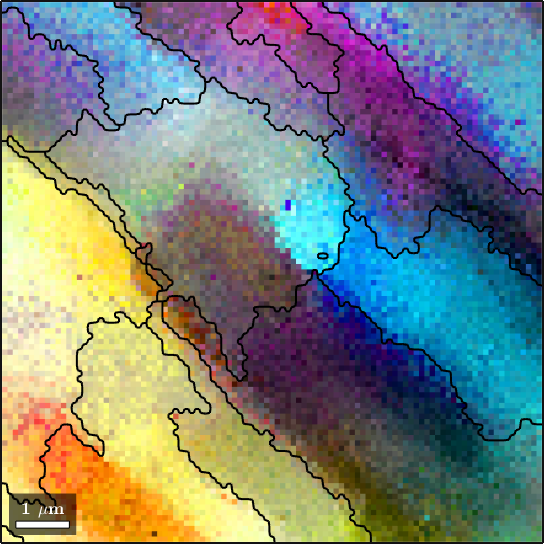
Markovian Clustering Algorithm
F = halfQuadraticFilter
F.alpha = 0.5
ebsd = smooth(ebsd,F)F =
halfQuadraticFilter with properties:
l1DataFit: 1
l1TV: 1
alpha: 1
iterMax: 1000
tol: 3.4907e-04
eps: 1.0000e-03
threshold: 0.2618
isHex: 0
F =
halfQuadraticFilter with properties:
l1DataFit: 1
l1TV: 1
alpha: 0.5000
iterMax: 1000
tol: 3.4907e-04
eps: 1.0000e-03
threshold: 0.2618
isHex: 0
ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
1 10201 (100%) Al LightSkyBlue m-3m
Properties: oldId, quality
Scan unit : um
X x Y x Z : [6, 16] x [4, 14] x [0, 0]
Normal vector: (0,0,1)grains = calcGrains(ebsd,'mcl',[1.24 50],'soft',[0.2 0.3]*degree)
grains = smooth(grains,5)
plot(ebsd,colorKey.orientation2color(ebsd.orientations))
hold on;plot(grains.boundary,'linewidth',2); hold offgrains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 285 10201 Al m-3m
boundary segments: 4107 (374 µm)
inner boundary segments: 16502 (1656 µm)
triple points: 423
Properties: meanRotation, GOS
grains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 285 10201 Al m-3m
boundary segments: 4107 (372 µm)
inner boundary segments: 16502 (1650 µm)
triple points: 423
Properties: meanRotation, GOS