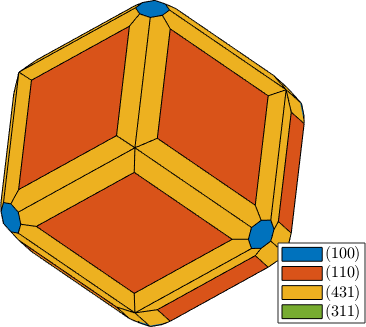
Crystal Shapes are used to visualize crystal orientations, twinning or lattice planes.
Simple crystal shapes
In the case of cubic or hexagonal materials the corresponding crystal are often represented as cubes or hexagons, where the faces correspond to the lattice planes {100} in the cubic case and {1,0,-1,0},{0,0,0,1} in the hexagonal case. Such simple crystal shapes may be created in MTEX with the commands
% import some hexagonal data
mtexdata titanium;
% define a simple hexagonal crystal shape
cS = crystalShape.hex(ebsd.CS)
% and plot it
close all
plot(cS,'faceAlpha',0.2)
drawNow(gcm,'final')ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 8100 (100%) Titanium (Alpha) LightSkyBlue 622 X||a, Y||b*, Z||c*
Properties: ci, grainid, iq, sem_signal
Scan unit : um
X x Y x Z : [0, 996] x [0, 998] x [0, 0]
Normal vector: (0,0,1)
cS = crystalShape
mineral: Titanium (Alpha) (622, X||a, Y||b*, Z||c*)
vertices: 12
faces: 8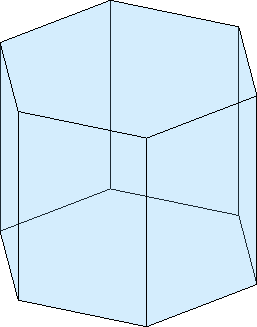
Internally, a crystal shape is represented as a list of faces which are bounded by a list of vertices cS.V and edges cS.E
cS.Vans = vector3d (y↑→x)
size: 12 x 1
x y z
-0.393614 0 -0.308331
-0.393614 0 0.308331
-0.196807 -0.34088 0.308331
-0.196807 0.34088 -0.308331
-0.196807 -0.34088 -0.308331
-0.196807 0.34088 0.308331
0.196807 -0.34088 0.308331
0.196807 0.34088 -0.308331
0.196807 -0.34088 -0.308331
0.196807 0.34088 0.308331
0.393614 0 -0.308331
0.393614 0 0.308331Using the commands plotInnerFace, plotInnerDirection, plot(cS,sS) and arrow3d we may plot internal lattice planes, directions or slip systems into the crystal shape
sS = [slipSystem.pyramidal2CA(ebsd.CS), ...
slipSystem.pyramidalA(ebsd.CS)]
plot(cS,'faceAlpha',0.2)
hold on
plot(cS,sS(2),'faceColor','blue')
plot(cS,sS(1),'faceColor','red')
hold off
drawNow(gcm,'final')sS = slipSystem (Titanium (Alpha))
size: 1 x 2
U V T W | H K I L CRSS
2 -1 -1 3 -2 1 1 2 1
2 -1 -1 0 0 1 -1 1 1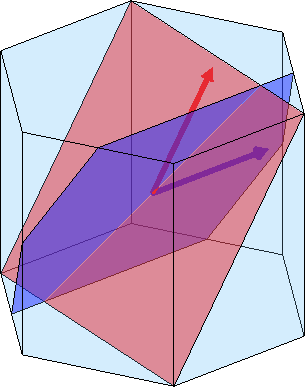
Calculating with crystal shapes
Crystal shapes are defined in crystal coordinates. Thus applying an orientation rotates them into specimen coordinates. This functionality can be used to visualize crystal orientations in EBSD maps
% plot an EBSD map
clf % clear current figure
plot(ebsd,ebsd.orientations)
hold on
scaling = 100; % scale the crystal shape to have a nice size
% plot at position (500,500) the orientation of the corresponding crystal
plot(500,500,50, ebsd(500,500).orientations * cS * scaling,'faceAlpha',0.5,'linewidth',2)
hold off
drawNow(gcm,'final')
As we have seen in the previous section we can apply several operations on crystal shapes. These include
-
factor * cSscales the crystal shape in size -
ori * cSrotates the crystal shape in the defined orientation -
[xy] + cSor[xyz] + cSshifts the crystal shape in the specified positions
At this point it comes into help that MTEX supports lists of crystal shapes, i.e., whenever one of the operations listed above includes a list (e.g. a list of orientations) the multiplication will yield a list of crystal shapes. Lets illustrate this
% compute some grains
grains = calcGrains(ebsd);
grains = smooth(grains,5);
% and plot them
cKey = ipfColorKey(grains);
color = cKey.orientation2color(grains.meanOrientation);
plot(grains,color,'FaceAlpha',0.5,'linewidth',2)
% find the big ones
isBig = grains.numPixel>50;
% define a list of crystal shape that is oriented as the grain mean
% orientation and scaled according to the grain area
cSGrains = grains(isBig).meanOrientation * cS * 0.7 * sqrt(grains(isBig).area);
% now we can plot these crystal shapes at the grain centers
hold on
plot(grains(isBig).centroid + cSGrains,'FaceColor',color(isBig,:),'FaceAlpha',0.7)
hold off
drawNow(gcm,'final')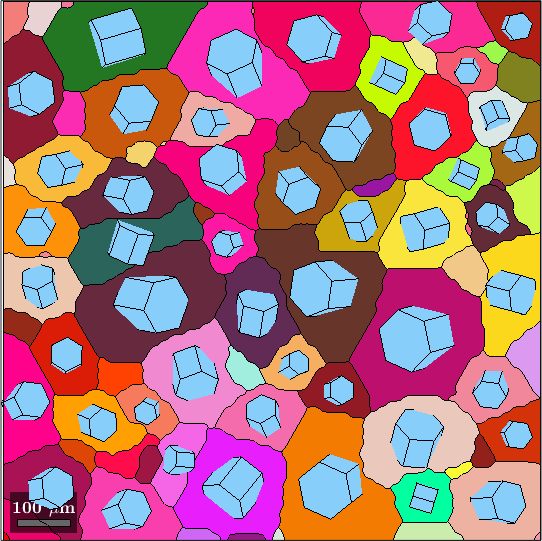
Plotting crystal shapes
The above can be accomplished a bit more directly and a bit more nice with
% plot a grain map
plot(grains,grains.meanOrientation,'figSize','large','faceAlpha',0.5,'linewidth',2)
%plot(grains.boundary,'figSize','large','linewidth',4)
% and on top for each large grain a crystal shape colored according to the
% grain orientation
hold on
plot(grains(isBig), 0.7*cS, 'FaceColor', color(isBig,:), ...
'linewidth',2,'FaceAlpha',0.7 )
hold off
drawNow(gcm,'final')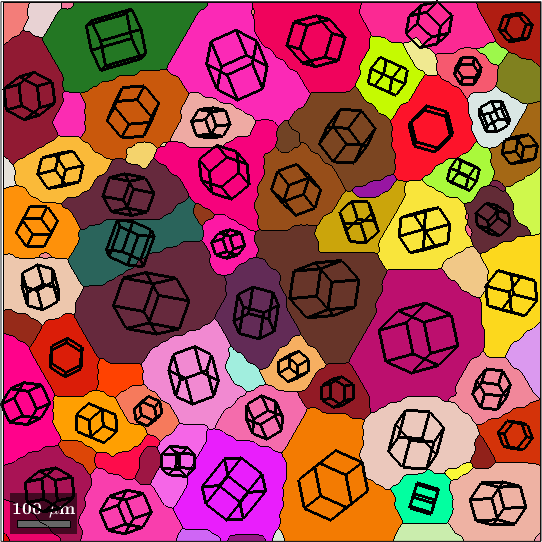
In the same way we may visualize grain orientations and grains size within pole figures
plotPDF(grains(isBig).meanOrientation,Miller({1,0,-1,0},{0,0,0,1},ebsd.CS),'contour')
plot(grains(isBig).meanOrientation,0.002*cSGrains,'add2all')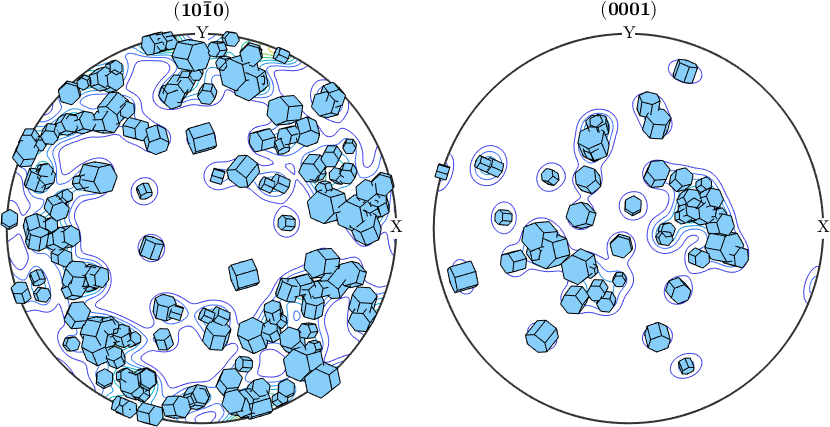
or even within ODF sections
% compute the odf
odf = calcDensity(ebsd.orientations);
% plot the odf in sigma sections
plotSection(odf,'sigma','contour')
% and on top of it the crystal shapes
plot(grains(isBig).meanOrientation,0.002*cSGrains,'add2all')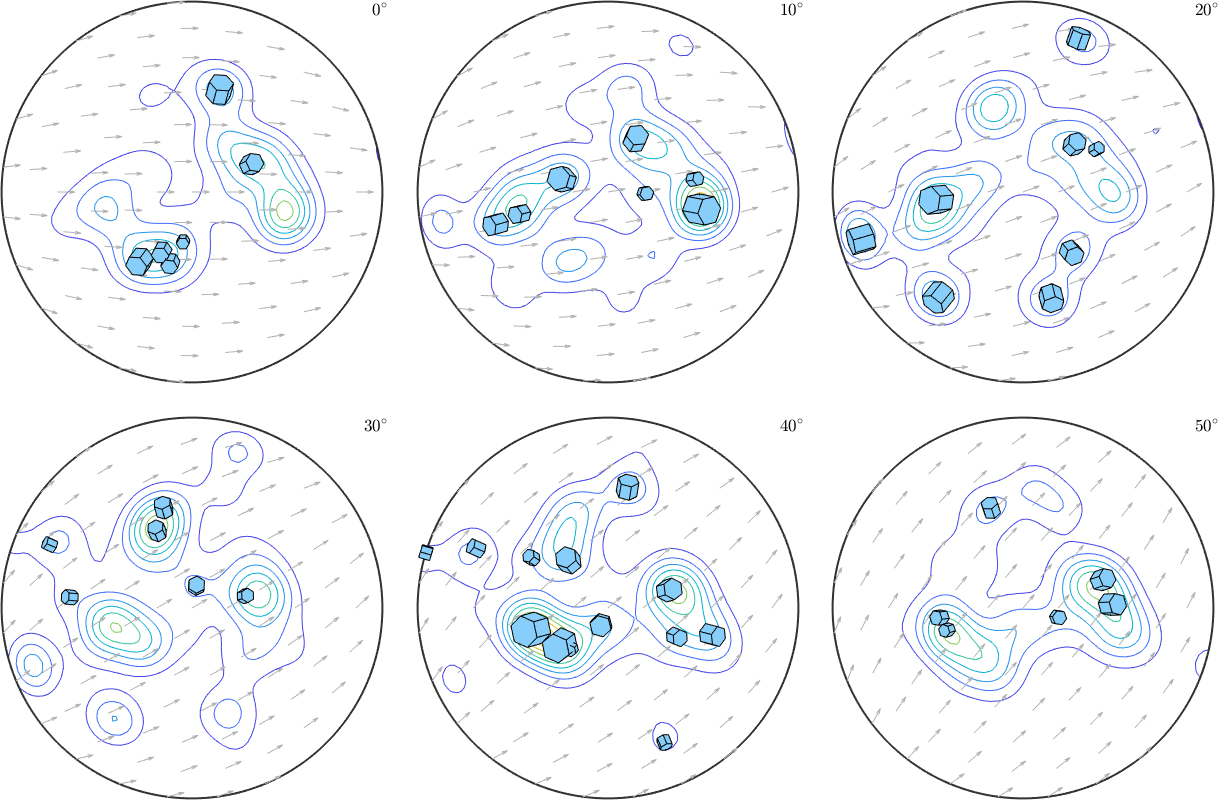
Twinning relationships
We may also you crystal shapes to illustrate twinning relation ships
% define some twinning misorientation
mori = orientation.byAxisAngle(Miller({1 0-1 0},ebsd.CS),34.9*degree)
% plot the crystal in ideal orientation
close all
plot(cS,'FaceAlpha',0.5)
% and on top of it in twinning orientation
hold on
plot(mori * cS *0.9,'FaceColor','orange')
hold off
view(45,20)
drawNow(gcm,'final')mori = misorientation (Titanium (Alpha) → Titanium (Alpha))
Bunge Euler angles in degree
phi1 Phi phi2
330 34.9 30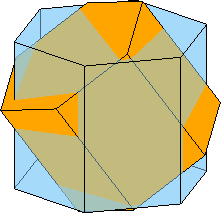
Defining complicated crystal shapes
For symmetries other then hexagonal or cubic one would like to have more complicated crystal shape representing the true appearance. To this end one has to include more faces into the representation and carefully adjust their distance to the origin.
Lets consider a quartz crystal.
cs = loadCIF('quartz')cs = crystalSymmetry
mineral : Quartz
symmetry : 321
elements : 6
a, b, c : 4.9, 4.9, 5.4
reference frame: X||a*, Y||b, Z||c*Its shape is mainly bounded by the following faces
m = Miller({1,0,-1,0},cs); % hexagonal prism
r = Miller({1,0,-1,1},cs); % positive rhomboedron, usally bigger then z
z = Miller({0,1,-1,1},cs); % negative rhomboedron
s1 = Miller({2,-1,-1,1},cs);% left tridiagonal bipyramid
s2 = Miller({1,1,-2,1},cs); % right tridiagonal bipyramid
x1 = Miller({6,-1,-5,1},cs);% left positive Trapezohedron
x2 = Miller({5,1,-6,1},cs); % right positive TrapezohedronIf we take only the first three faces we end up with
N = [m,r,z];
cS = crystalShape(N)
plot(cS)cS = crystalShape
mineral: Quartz (321, X||a*, Y||b, Z||c*)
vertices: 8
faces: 18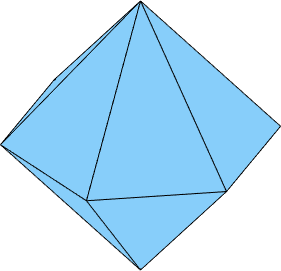
i.e. we see only the positive and negative rhododendrons, but the hexagonal prism are to far away from the origin to cut the shape. We may decrease the distance, by multiplying the corresponding normal with a factor larger then 1.
N = [2*m,r,z];
cS = crystalShape(N);
plot(cS,'colored')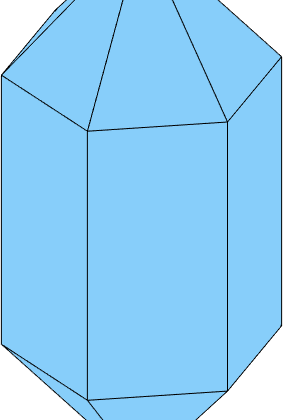
Next in a typical Quartz crystal the negative rhododendron is a bit smaller then the positive rhododendron. Lets correct for this.
% collect the face normal with the right scaling
N = [2*m,r,0.9*z];
cS = crystalShape(N);
plot(cS,'colored')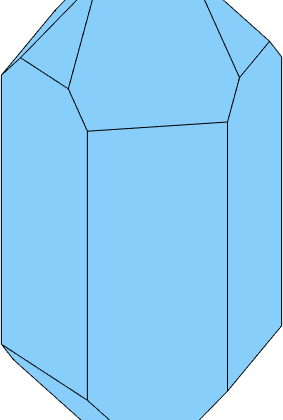
Finally, we add the tridiagonal bipyramid and the positive Trapezohedron
% collect the face normal with the right scaling
N = [2*m,r,0.9*z,0.7*s1,0.3*x1];
cS = crystalShape(N);
plot(cS,'colored')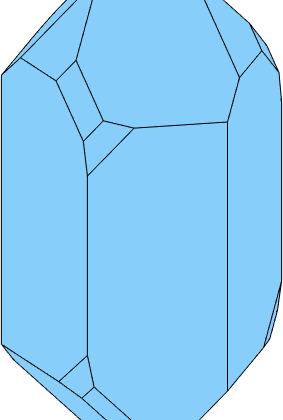
Defining complicated crystals more simple
We see that defining a complicated crystal shape is a tedious work. To this end MTEX allows to model the shape with a habitus and a extension parameter. This approach has been developed by J. Enderlein in A package for displaying crystal morphology. Mathematical Journal, 7(1), 1997. The two parameters are used to model the distance of a face from the origin. Setting all parameters to one we obtain
% take the face normals unscaled
N = [m,r,z,s2,x2];
habitus = 1;
extension = [1 1 1];
cS = crystalShape(N,habitus,extension);
plot(cS,'colored')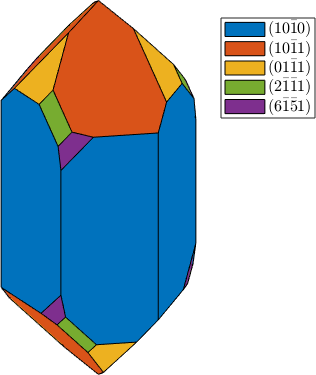
The scale parameter models the inverse extension of the crystal in each dimension. In order to make the crystal a bit longer and the negative rhododendrons smaller we could do
extension = [1 1.2 1.1];
cS = crystalShape(N,habitus,extension);
plot(cS,'colored')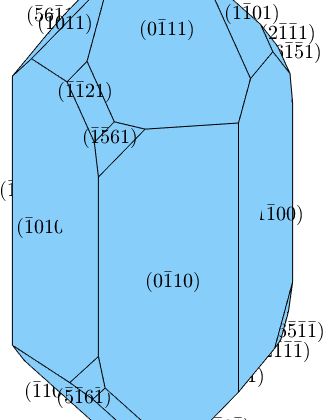
Next the habitus parameter describes how close faces with mixed hkl are to the origin. If we increase the habitus parameter the trapezohedron and the bipyramid become more and more dominant
habitus = 1.1;
cS = crystalShape(N,habitus,extension);
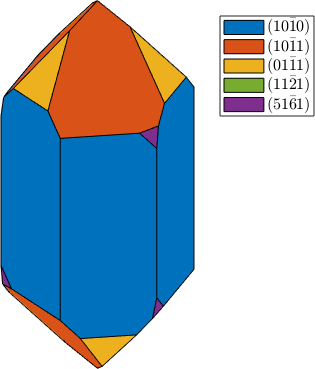
plot(cS,'colored'), snapnow
habitus = 1.2;
cS = crystalShape(N,habitus,extension);
plot(cS,'colored'), snapnow
habitus = 1.3;
cS = crystalShape(N,habitus,extension);
plot(cS,'colored')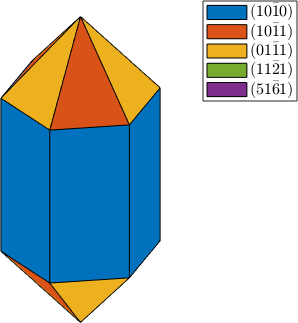
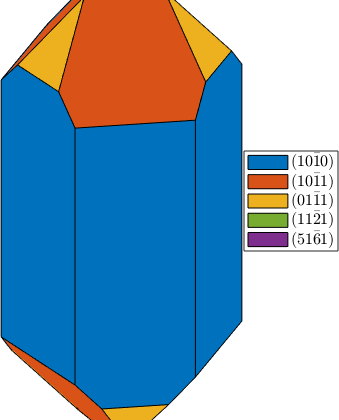
Select faces
A specific face of the crystal shape may be selected by its normal vector
plot(cS)
hold on
plot(cS(Miller(0,-1,1,0,cs)),'FaceColor','DarkRed')
hold off
% zoom a bit out to fit the screen
camzoom(0.7)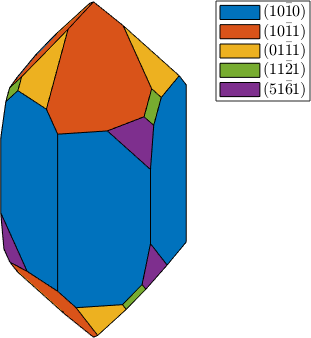
Gallery of hardcoded crystal shapes
plot(crystalShape.olivine,'colored')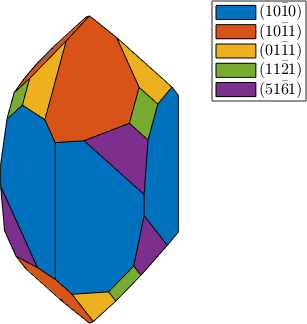
plot(crystalShape.garnet,'colored')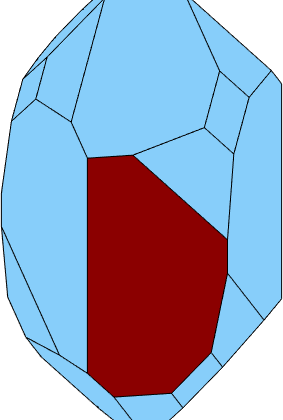
plot(crystalShape.topaz,'colored')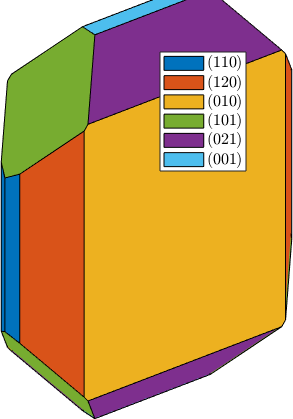
plot(crystalShape.plagioclase,'colored')