How to analyze slip transmission at grain boundaries
Import Titanium data
From Mercier D. - MTEX 2016 Workshop - TU Chemnitz (Germany) Calculation and plot on GBs of m' parameter Dataset from Mercier D. - cp-Ti (alpha phase - hcp)
mtexdata titanium
% compute grains
[grains, ebsd.grainId] = calcGrains(ebsd('indexed'));
% make them a bit nicer
grains = smooth(grains);
% extract inner phase grain boundaries
gB = grains.boundary('indexed');
plot(ebsd,ebsd.orientations)
hold on
plot(grains.boundary)
hold offebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 8100 (100%) Titanium (Alpha) LightSkyBlue 622 X||a, Y||b*, Z||c*
Properties: ci, grainid, iq, sem_signal
Scan unit : um
X x Y x Z : [0, 996] x [0, 998] x [0, 0]
Normal vector: (0,0,1)
Schmid Factor
% consider Basal slip
sSBasal = slipSystem.basal(ebsd.CS)
% and all symmetrically equivalent variants
sSBasal = sSBasal.symmetrise;
% compute Schmid factor for all slip systems
SF = sSBasal.SchmidFactor(inv(grains.meanOrientation) * xvector);
% find the maximum Schmidt factor
[SF,id] = max(SF,[],2);
% and plot it for each grain
plot(grains,SF)
mtexColorbarsSBasal = slipSystem (Titanium (Alpha))
U V T W | H K I L CRSS
1 1 -2 0 0 0 0 1 1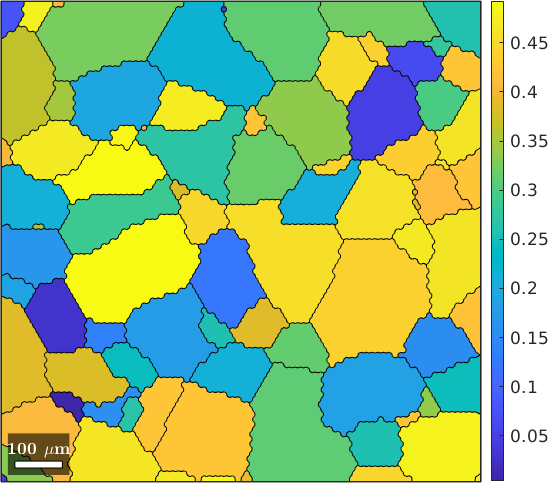
The variable id contains now for each grain the id of the slip system with the largest Schmidt factor. In order to visualize it, we first rotate for each grain the slip system with largest Schmid factor in specimen coordinates
sSGrain = grains.meanOrientation .* sSBasal(id)
% and then plot the plane normal and the Burgers vectors into the centers
% of the grains
hold on
quiver(grains,sSGrain.trace,'displayName','slip plane')
hold on
quiver(grains,sSGrain.b,'displayName','slip direction','project2plane')
hold off
legend showsSGrain = slipSystem (y↑→x)
CRSS: 1
size: 85 x 1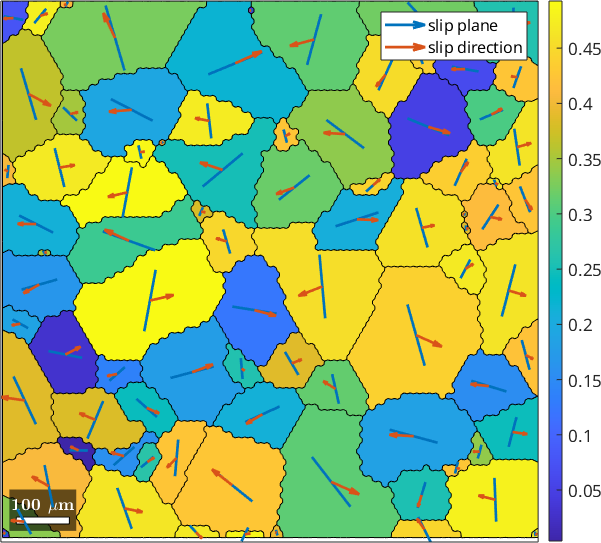
We may also analyze the distribution of the slip directions in a pole figure plot
plot(sSGrain.b)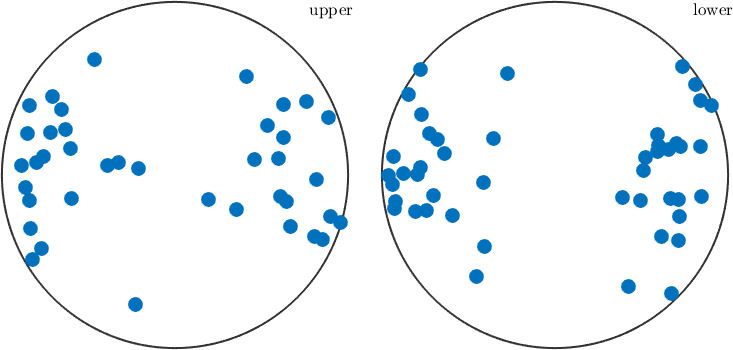
The same as a contour plot. We see a clear trend in east/west direction.
plot(sSGrain.b,'contourf')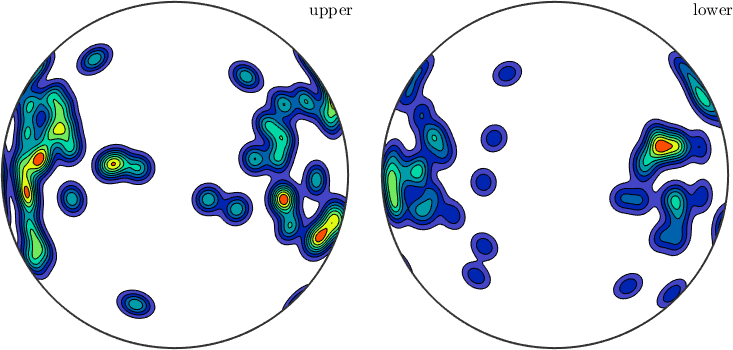
Resolved shear stress
We could do the same as above with a more general stress tensor. However, the result is a bit different.
% more general syntax for the same stress tensor
sigma = stressTensor.uniaxial(xvector);
% compute Schmid factor for all slip systems
SF = sSBasal.SchmidFactor(inv(grains.meanOrientation) * sigma);
% find the maximum Schmidt factor
[SF,id] = max(SF,[],2);
% plot the Schmid factor
plot(grains,SF)
mtexColorbar
% active slip system in specimen coordinates
sSGrain = grains.meanOrientation .* sSBasal(id)
% and plot then the plane normal and the Burgers vectors into the centers
% of the grains
hold on
quiver(grains,sSGrain.trace,'displayName','slip plane')
hold on
quiver(grains,sSGrain.b,'displayName','slip direction','project2plane')
hold off
legend showsSGrain = slipSystem (y↑→x)
CRSS: 1
size: 85 x 1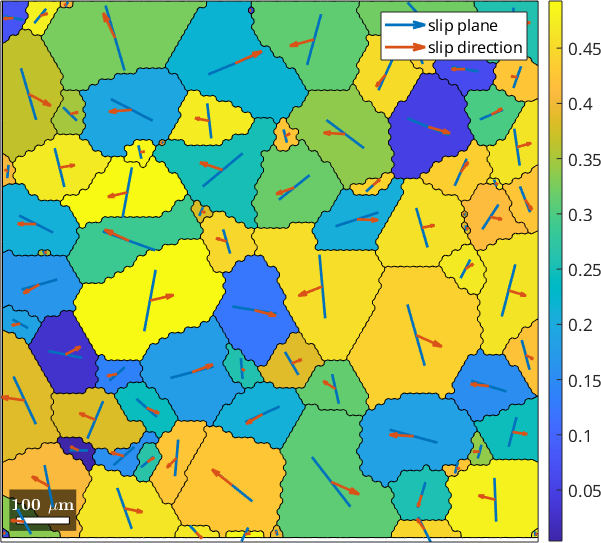
Compatibility of slip systems
Next, we want to analyze, how much geometrically compatible the slip systems with highest Schmid factor are at the grain boundaries
% some background
plot(grains,'FaceColor',0.8*[1 1 1],'figSize','large')
% compute m'
id = gB.grainId;
mP = mPrime(sSGrain(id(:,1)),sSGrain(id(:,2)));
% plot m' along the grain boundaries
hold on
plot(gB,mP,'linewidth',3)
mtexColorbar
% and plot then the plane normal and the Burgers vectors into the centers
% of the grains
hold on
quiver(grains,sSGrain.trace,'displayName','slip plane')
hold on
quiver(grains,sSGrain.b,'displayName','slip direction','project2plane')
hold off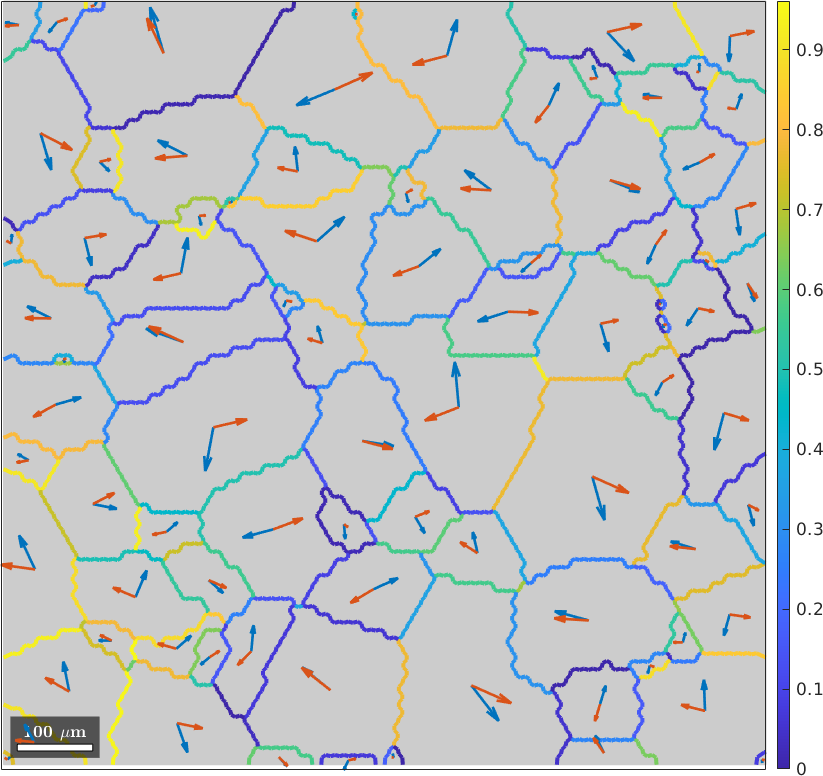
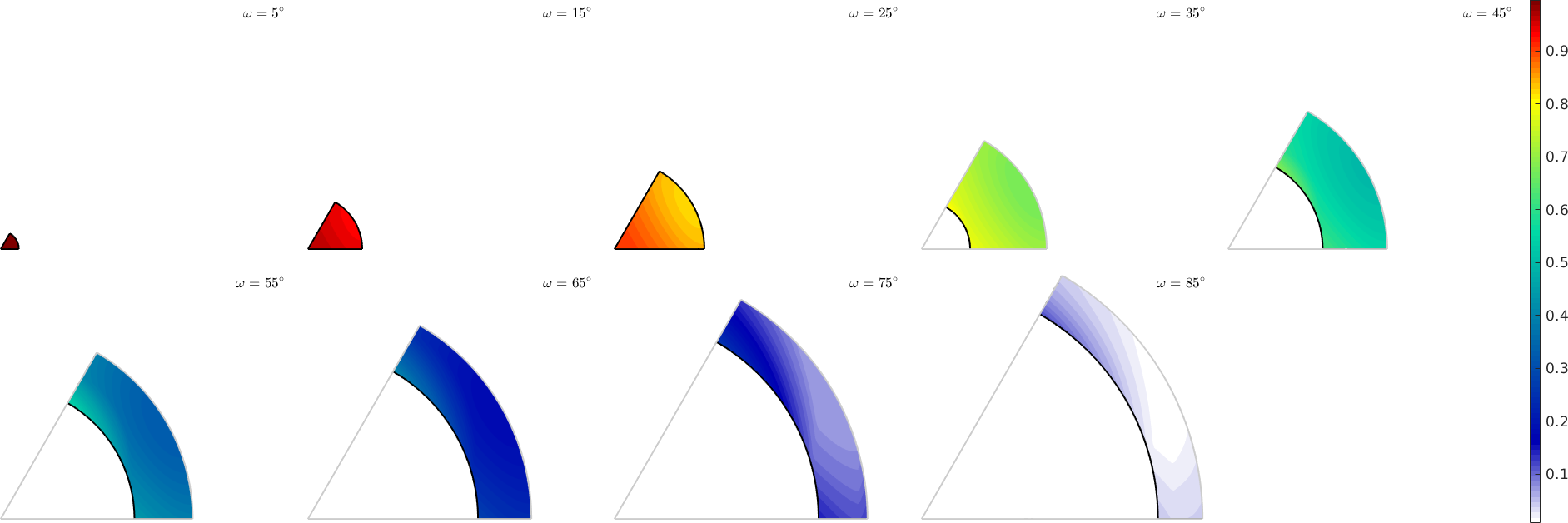
Plot m' in misorientation space
Since m' depends on the misorientation only why may plot it directly in misorientation space
% set up an axis angle plot
sP = axisAngleSections(sSBasal.CS,sSBasal.CS);
% generate a grid of misorientations
moriGrid = sP.makeGrid;
% compute mPrime for all misorientations
sSBasal = slipSystem.basal(ebsd.CS);
mP = max(mPrime(sSBasal,moriGrid*sSBasal.symmetrise),[],2);
% plot mPrime
sP.plot(mP,'smooth')
mtexColorbar