We will prepare some data to evaluate grain dispersion axes.
mtexdata forsterite
[grains,ebsd.grainId] = ebsd.calcGrains;
% just use the larger grains of forsterite
ebsd(grains(grains.numPixel< 100))='notIndexed';
ebsd({'e' 'd'})='notIndexed';
% lets also ignore inclusions for a nicer plotting experience
ebsd(grains(grains.isInclusion))=[];
[grains,ebsd.grainId, ebsd.mis2mean] = ebsd.calcGrains;ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 58485 (24%) notIndexed
1 152345 (62%) Forsterite LightSkyBlue mmm
2 26058 (11%) Enstatite DarkSeaGreen mmm
3 9064 (3.7%) Diopside Goldenrod 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [0, 36550] x [0, 16750] x [0, 0]
Normal vector: (0,0,1)We colorize axes of the misorientation to the grain mean orientation in specimen coordinates
ck = axisAngleColorKey(ebsd('f').CS);
ck.oriRef=grains('id',ebsd('f').grainId).meanOrientation;
plot(ebsd('f'), ck.orientation2color(ebsd('f').orientations))
hold on
plot(grains.boundary)
hold on
plot(grains,'FaceAlpha',0.3)
hold off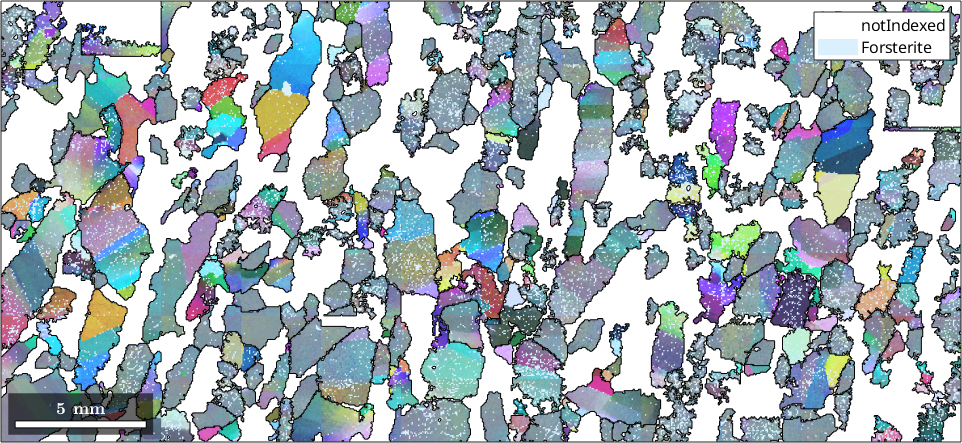
Visualizing dispersion of orientations via directions
% First, we will inspect a selected grain
grain_selected = grains(5095, 7803);
hold on
plot(grain_selected.boundary,'linewidth',3,'linecolor','b')
hold off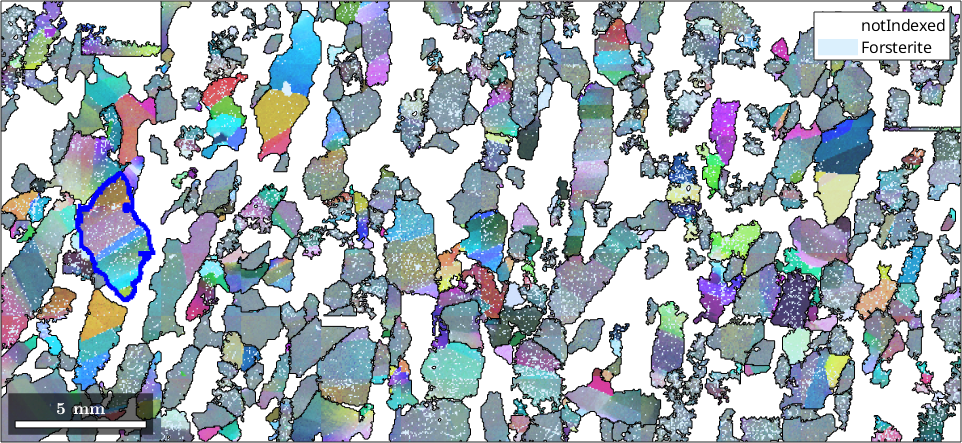
and examine the spread of orientations in terms of its pole figure. In order to do so, we can define a grid of crystal direction and compute the corresponding specimen directions for each orientation within the grain.
% Let's define a grid of directions
s2G = equispacedS2Grid('resolution',15*degree);
s2G = Miller(s2G,ebsd('f').CS)
% use the orientations of points belonging to the grain
o = ebsd(grain_selected).orientations;
% and compute the corresponding specimen directions
d = o .* s2G;
% and plot them
plot(d,'MarkerSize',3,'upper')
% We can observe, that certain grid points are smeared out more than otherss2G = Miller (Forsterite)
size: 1 x 184
resolution: 15°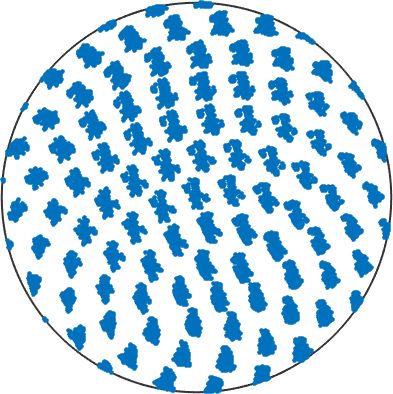
%Next, we compute the mean angular deviation for each group of grid points
vd = mean(angle(mean(d),d));
% and plot those
plot(d,repmat(vd,length(o),1)/degree,'MarkerSize',3)
mtexColorbar('title','avergage pole dispersion')
% and we can ask which grid point is the one with the smallest dispersion
[~,id_min]=min(vd);
disp_ax_grid = grain_selected.meanOrientation .* s2G(id_min);
annotate(disp_ax_grid)
annotate(disp_ax_grid,'plane','linestyle','--','linewidth',2)
% While we might have guessed the result by eye, it is not too satisfying since
% the direction of the estimated dispersion axis will always be located
% on a grid point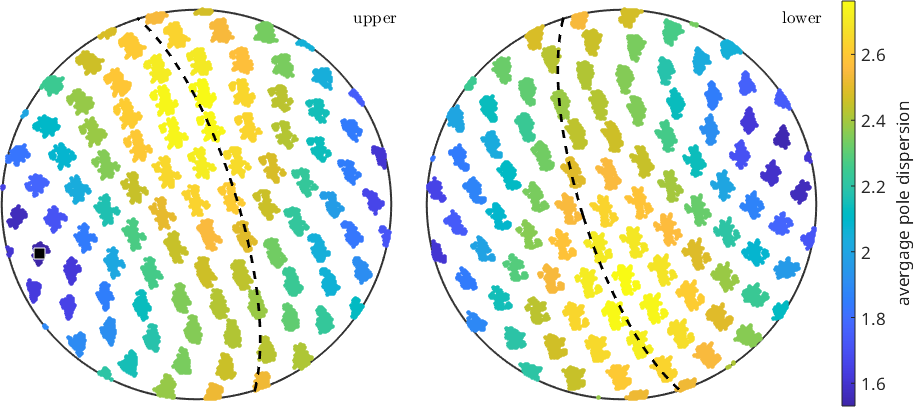
If we assume, the orientations are dispersed along one single axis, we can fit an orientation fibre fibre
% This can be accomplished by |fibre.fit|
fib = fibre.fit(o,'local')
% the fibre has an axis in specimen coordinates |fib.r| and in crystal
% coordinates |fib.h|.
fib.r
fib.h
% and we can visualize those also in our previous plot
annotate(fib.r,'MarkerFaceColor','r')
annotate(fib.r,'plane','linestyle','-.','linewidth',2,'lineColor','r')fib = fibre (Forsterite → y↑→x)
h || r: (-1-112) || (-12,-5,3)
o1 → o2: (12.8°,44.7°,280.8°) → (12.8°,44.7°,280.8°)
ans = vector3d (y↑→x)
x y z
-0.89864 -0.376589 0.225003
ans = Miller (Forsterite)
h k l
-1.0362 -9.5489 1.6628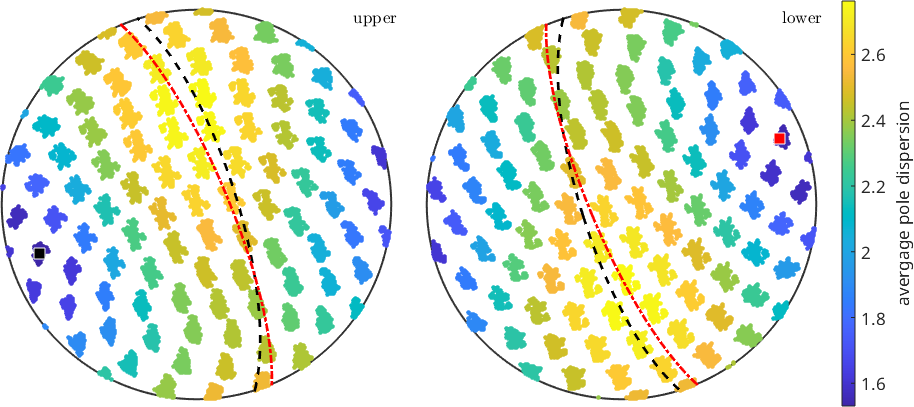
We can also inspect in orientation space, how well the fibre fits the orientations of the grain
% The angle between each orientation and the fibre gives a measure how well
% it is fitted by the fibre
fd = angle(fib,o)/degree;
plot(o,fd)
xlim([0 30]); ylim([20 70]); zlim([80 120])
grid minor
hold on
plot(fib,'linewidth',2)
hold off
nextAxis
% we can also inspect the distance of each orientation within the grains
% to the fitted fibre with the grains
plot(ebsd(grain_selected),fd)
mtexColorbar('title', 'distance from fibre')plot 2000 random orientations out of 2826 given orientations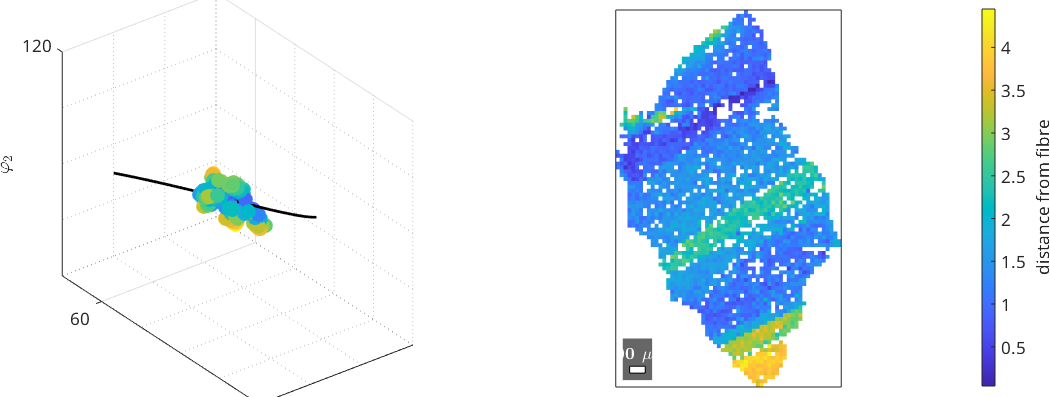
TODO: use eigenvalues of fibre.fit to give measure of "fibryness" [fib, lambda] = fibre.fit(o,'local') lambda(2)/lambda(3)
Bulk evaluation
We can fit a fibre for each grain and write out the axes in crystal as well as in specimen coordinates
%ids = grains('f').id;
%clear fib_axSC fib_axCC
%for i = 1:length(ids)
% o = ebsd(grains('id',ids(i))).orientations;
% fib = fibre.fit(o);
% fib_axCC(i) = fib.h;
% fib_axSC(i) = fib.r;
%end
% plot fibre axes in specimen coordinates
%opts= {'contourf','antipodal','halfwidth', 10*degree,'contours',[1:10]};
%plot(fib_axSC,opts{:})
%nextAxis
% plot fibre axes in crystal coordinates
%plot(fib_axCC,opts{:},'fundamentalRegion')
%mtexColorbar
% Now we can start to wonder whether the distrubtion of fibre axes relates
% to e.g. the kinematic during deformation of the sample.