A quick guide to grain boundary analysis
Grain boundaries generation
To work with grain boundaries we need some ebsd data and have to detect grains within the data set.
% load some example data
mtexdata twins
% detect grains
[grains,ebsd.grainId,ebsd.mis2mean] = calcGrains(ebsd('indexed'))
% smooth them
grains = grains.smooth
% visualize the grains
plot(grains,grains.meanOrientation)ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 46 (0.2%) notIndexed
1 22833 (100%) Magnesium LightSkyBlue 6/mmm X||a*, Y||b, Z||c*
Properties: bands, bc, bs, error, mad
Scan unit : um
X x Y x Z : [0, 50] x [0, 41] x [0, 0]
Normal vector: (0,0,1)
grains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 121 22833 Magnesium 6/mmm X||a*, Y||b, Z||c*
boundary segments: 4416 (1154 µm)
inner boundary segments: 4 (1.2 µm)
triple points: 114
Properties: meanRotation, GOS
ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 46 (0.2%) notIndexed
1 22833 (100%) Magnesium LightSkyBlue 6/mmm X||a*, Y||b, Z||c*
Properties: bands, bc, bs, error, mad, grainId, mis2mean
Scan unit : um
X x Y x Z : [0, 50] x [0, 41] x [0, 0]
Normal vector: (0,0,1)
ebsd = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
0 46 (0.2%) notIndexed
1 22833 (100%) Magnesium LightSkyBlue 6/mmm X||a*, Y||b, Z||c*
Properties: bands, bc, bs, error, mad, grainId, mis2mean
Scan unit : um
X x Y x Z : [0, 50] x [0, 41] x [0, 0]
Normal vector: (0,0,1)
grains = grain2d (y↑→x)
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 121 22833 Magnesium 6/mmm X||a*, Y||b, Z||c*
boundary segments: 4416 (1022 µm)
inner boundary segments: 4 (0.99 µm)
triple points: 114
Properties: meanRotation, GOS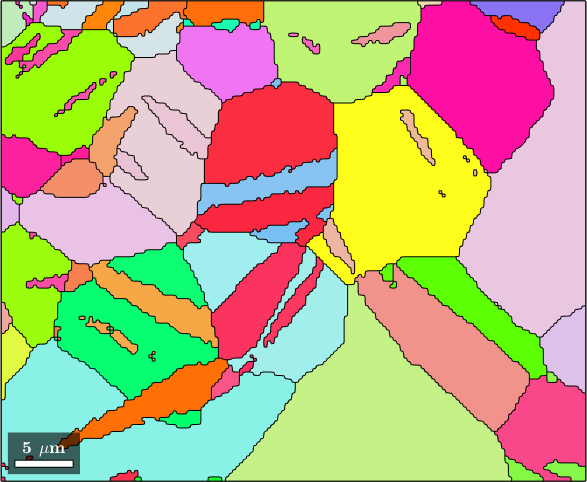
Now we can extract from the grains its boundary and save it to a separate variable
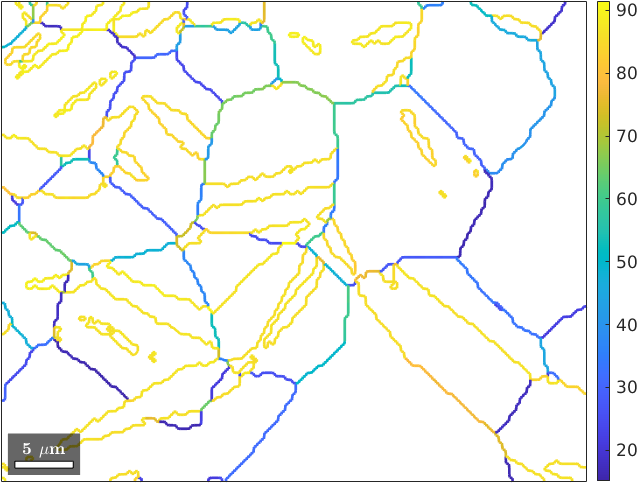
gB = grains.boundarygB = grainBoundary
Segments length mineral 1 mineral 2
1197 184 µm notIndexed Magnesium
3219 837 µm Magnesium MagnesiumThe output tells us that we have 3219 Magnesium to Magnesium boundary segments and 606 boundary segments where the grains are cut by the scanning boundary. To restrict the grain boundaries to a specific phase transition you shall do
gB_MgMg = gB('Magnesium','Magnesium')gB_MgMg = grainBoundary
Segments length mineral 1 mineral 2
3219 837 µm Magnesium MagnesiumProperties of grain boundaries
A variable of type grain boundary contains the following properties
- misorientation
- direction
- segLength
These can be used to colorize the grain boundaries. By the following command we plot the grain boundaries colorized by the misorientation angle
plot(gB_MgMg,gB_MgMg.misorientation.angle./degree,'linewidth',2)
mtexColorbar