sort orientations into clusters
Syntax
[c,center] = doHCluster(ori,'numCluster',n)
[c,center] = doHCluster(ori,'maxAngle',omega)Input
| ori | orientation |
| n | number of clusters |
| omega | maximum angle |
Output
| c | list of clusters |
| center | center of the clusters |
Example
% generate orientation clustered around 5 centers
cs = crystalSymmetry('m-3m');
center = orientation.rand(5,cs);
odf = unimodalODF(center,'halfwidth',5*degree)
ori = odf.discreteSample(3000);odf = SO3FunRBF (m-3m → y↑→x)
multimodal components
kernel: de la Vallee Poussin, halfwidth 5°
center: 5 orientations
Bunge Euler angles in degree
phi1 Phi phi2 weight
156.958 109.836 223.608 0.2
9.33344 126.207 190.491 0.2
197.878 76.1995 48.4488 0.2
156.716 113.621 184.888 0.2
151.332 117.797 66.3984 0.2% find the clusters and its centers
tic; [c,centerRec] = calcCluster(ori,'method','hierarchical','numCluster',5); tocElapsed time is 3.419015 seconds.% visualize result
oR = fundamentalRegion(cs)
plot(oR)oR = orientationRegion
crystal symmetry: 432
max angle: 62.7994°
face normales: 14
vertices: 24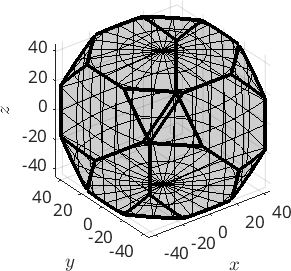
hold on
plot(ori,ind2color(c))
caxis([1,5])
plot(center,'MarkerSize',10,'MarkerFaceColor','k','MarkerEdgeColor','k')
plot(centerRec,'MarkerSize',10,'MarkerFaceColor','r','MarkerEdgeColor','k')
hold offplot 2000 random orientations out of 3000 given orientations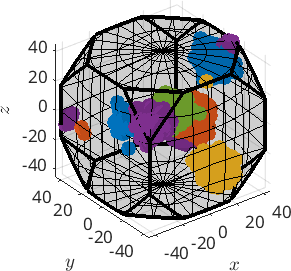
%check the accuracy of the recomputed centers
min(angle_outer(center,centerRec)./degree)ans =
0.30315 0.32977 0.23661 0.54501 0.15798