Electron Backscatter Diffraction Data (The Class EBSD)
This section describes the class EBSD and gives an overview over the functionality that MTEX offers to analyze EBSD data.
| On this page ... |
| Class Description |
| Import of EBSD Data |
| Plotting EBSD Data |
| Modify EBSD Data |
| Calculate an ODF from EBSD Data |
| Simulate EBSD Data |
| Demo |
| Complete Function list |
Class Description
The following mindmap might give a basic idea about EBSD data analyis in MTEX, with the ability of grain modelling for spatial data. It offers various way of interpreting individual orientation measurements, i.e. in terms of quantifying macro-,micro- and mesotexture.

Import of EBSD Data
The most comfortable way to import EBSD data into MTEX is to use the import wizard, which can be started by the command
import_wizard('ebsd')
If the dataset is in a format supported by MTEX, the import wizard generates a script which imports the data. More information about the import wizard and a list of supported file formats can be found ImportEBSDData.html,here. A typical script generated by the import wizard looks a follows.
cs = crystalSymmetry('m-3m','mineral','Fe'); % crystal symmetry % file names fname = fullfile(mtexDataPath,'EBSD','85_829grad_07_09_06.txt'); % load data ebsd = EBSD.load(fname,'CS',cs,... 'Bunge','ignorePhase',[0 2],... 'ColumnNames', { 'Phase' 'x' 'y' 'Euler 1' 'Euler 2' 'Euler 3'},... 'Columns', [2 3 4 5 6 7])
ebsd = EBSD
Phase Orientations Mineral Color Symmetry Crystal reference frame
1 48184 (100%) Fe light blue m-3m
Properties: x, y
Scan unit : um
Plotting EBSD Data
EBSD data are plotted using the command EBSD.plot.html,plot. It assigns a color to each orientation and plots a map of these colors. There are several options to specify the way the colors are assigned.
ipfKey = ipfColorKey(ebsd); plot(ebsd,ipfKey.orientation2color(ebsd.orientations))
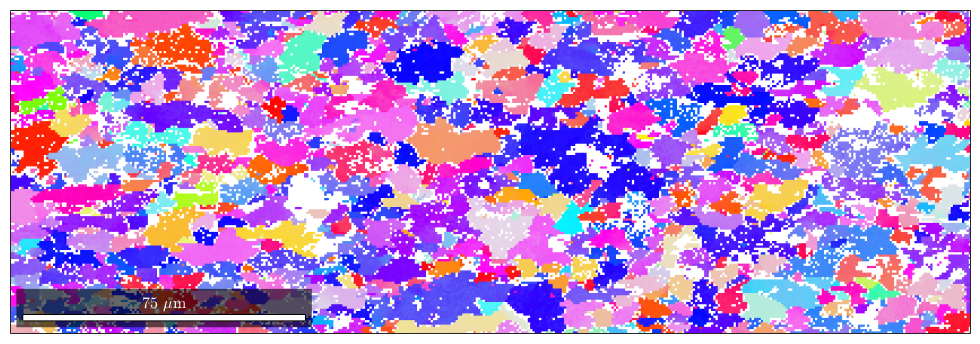
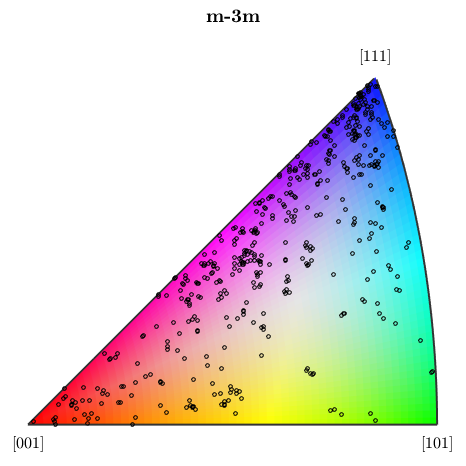
In order to understand the color coding, one can plot the coloring of the corresponding inverse pole figure via
plot(ipfKey) hold on plotIPDF(ebsd('Fe').orientations,xvector,'markerSize',3,'points',500,'marker','o','markerfacecolor','none','markeredgecolor','k') hold off
I'm plotting 500 random orientations out of 48184 given orientations
Modify EBSD Data
MTEX offers a lot of operations to analyze and manipulate EBSD data, e.g.
- plot pole figures of EBSD data
- rotate EBSD data
- find outliers
- remove specific measurements
- combine EBSD data from several measurements
- compute an ODF
An exhaustive introduction how to analyze and modify EBSD data can be found here
Calculate an ODF from EBSD Data
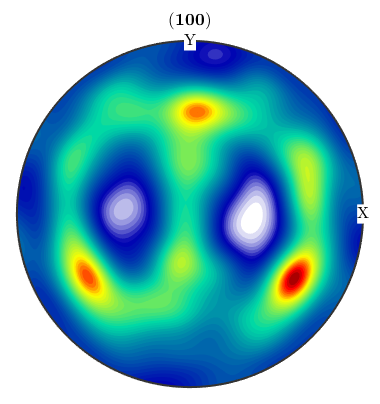
The command calcODF performs an ODF calculation from EBSD data using kernel density estimation EBSD data. For a precise explanation of the algorithm and the available options please look at here.
odf = calcODF(ebsd('Fe').orientations,'halfwidth',10*degree) plotPDF(odf,Miller(1,0,0,odf.CS),'antipodal')
odf = ODF
crystal symmetry : Fe (m-3m)
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
Simulate EBSD Data
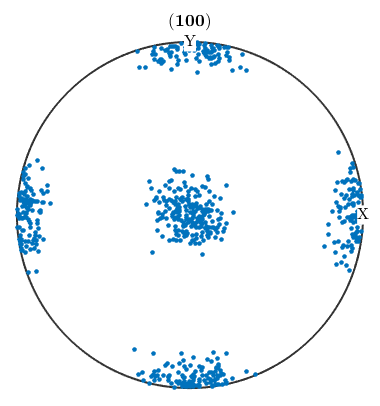
Simulating EBSD data from a given ODF is useful to analyze the stability of the ODF estimation process. There is an example demonstrating how to determine the number of individual orientation measurements to estimate the ODF up to a given error. The MTEX command to simulate EBSD data is calcEBSD, e.g.
ori = calcOrientations(unimodalODF(orientation.id(cs)),500) plotPDF(ori,Miller(1,0,0,cs),'antipodal','MarkerSize',3)
ori = orientation size: 500 x 1 crystal symmetry : Fe (m-3m) specimen symmetry: 1 I'm plotting 208 random orientations out of 500 given orientations You can specify the the number points by the option "points". The option "all" ensures that all data are plotted
Demo
For a more exhaustive description of the EBSD class have a look at the EBSD demo!
Complete Function list
| *EBSD* is the low level constructor for an *EBSD* object representing EBSD | |
| data. For importing real world data you might want to use the predefined | |
| <ImportEBSDData.html EBSD interfaces>. You can also simulate EBSD data | |
| from an ODF by the command <ODF.calcEBSD.html calcEBSD>. | |
| Syntax | |
| ebsd = EBSD(rotations,phases,CSList) | |
| Input | |
| orientations | @orientation |
| CS | crystal / specimen @symmetry |
| Options | |
| phase | specifing the phase of the EBSD object |
| options | struct with fields holding properties for each orientation |
| xy | spatial coordinates n x 2, where n is the number of input orientations |
| unitCell | for internal use |
| See also | |
| ODF/calcEBSD EBSD/calcODF EBSD/load |
| DocHelp 0.1 beta |