ODF Estimation from EBSD data
How to estimate an ODF from single orientation measurements.
| On this page ... |
| ODF Estimation |
| Automatic halfwidth selection |
| Effect of halfwidth selection |
ODF Estimation
These EBSD datasets consist of two phases, Iron, and Magnesium. The ODF of the Iron phase is computed by the command
odf = calcODF(ebsd('fo').orientations)
odf = ODF
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
The function calcODF implements the ODF estimation from EBSD data in MTEX. The underlying statistical method is called kernel density estimation,
which can be seen as a generalized histogram. To be more precise, let's 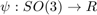 be a radially symmetric, unimodal model ODF. Then the kernel density estimator for the individual orientation data
be a radially symmetric, unimodal model ODF. Then the kernel density estimator for the individual orientation data  is defined as
is defined as
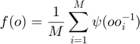
The choice of the model ODF 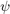 and in particular its halfwidth has a great impact in the resulting ODF. If no halfwidth is specified the default halfwidth
of 10 degrees is selected.
and in particular its halfwidth has a great impact in the resulting ODF. If no halfwidth is specified the default halfwidth
of 10 degrees is selected.
Automatic halfwidth selection
MTEX includes an automatic halfwidth selection algorithm which is called by the command calcKernel. To work properly, this algorithm needs spatially independent EBSD data as in the case of this dataset of very rough EBSD measurements (only one measurement per grain).
% try to compute an optimal kernel psi = calcKernel(ebsd('fo').orientations)
psi = deLaValleePoussinKernel bandwidth: 98 halfwidth: 2.3°
In the above example, the EBSD measurements are spatial dependent and the resulting halfwidth is too small. To avoid this problem we have to perform grain reconstruction first and then estimate the halfwidth from the grains.
% grains reconstruction grains = calcGrains(ebsd); % correct for to small grains grains = grains(grains.grainSize>5); % compute optimal halfwidth from the meanorientations of grains psi = calcKernel(grains('fo').meanOrientation) % compute the ODF with the kernel psi odf = calcODF(ebsd('fo').orientations,'kernel',psi)
psi = deLaValleePoussinKernel
bandwidth: 41
halfwidth: 6°
odf = ODF
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
Harmonic portion:
degree: 41
weight: 1
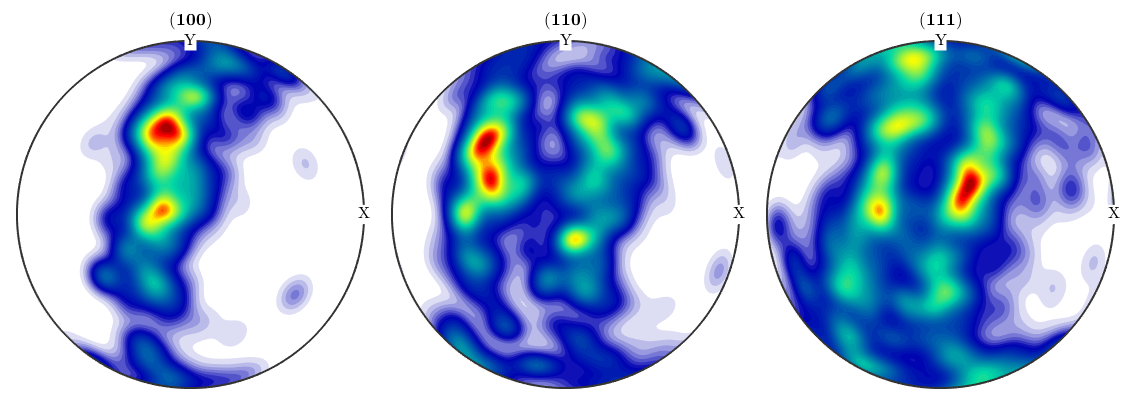
Once an ODF is estimated all the functionality MTEX offers for ODF analysis and ODF visualization is available.
h = [Miller(1,0,0,odf.CS),Miller(1,1,0,odf.CS),Miller(1,1,1,odf.CS)]; plotPDF(odf,h,'antipodal','silent')
Effect of halfwidth selection
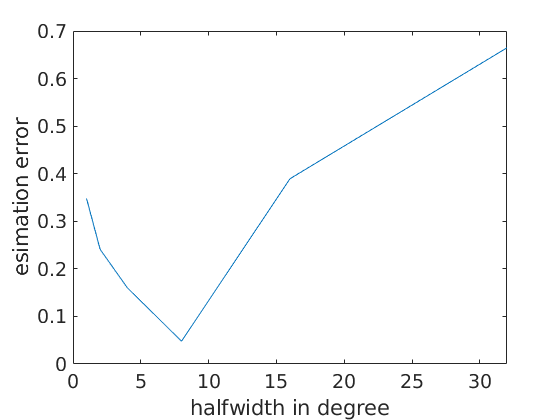
As mentioned above a proper halfwidth selection is crucial for ODF estimation. The following simple numerical experiment illustrates the dependency between the kernel halfwidth and the estimated error.
Let's start with a model ODF and simulate some individual orientation data.
modelODF = fibreODF(Miller(1,1,1,crystalSymmetry('cubic')),xvector);
ori = calcOrientations(modelODF,10000)ori = orientation size: 10000 x 1 crystal symmetry : m-3m specimen symmetry: 1
Next we define a list of kernel halfwidth ,
hw = [1*degree, 2*degree, 4*degree, 8*degree, 16*degree, 32*degree];
estimate for each halfwidth an ODF and compare it to the original ODF.
e = zeros(size(hw)); for i = 1:length(hw) odf = calcODF(ori,'halfwidth',hw(i),'silent'); e(i) = calcError(modelODF, odf); end
After visualizing the estimation error we observe that its value is large either if we choose a very small or a very large halfwidth. In this specific example, the optimal halfwidth seems to be about 4 degrees.
close all plot(hw/degree,e) xlabel('halfwidth in degree') ylabel('esimation error')
| DocHelp 0.1 beta |