MTEX - Analysis of EBSD Data
Analysis of single orientation measurement.
Specify Crystal and Specimen Symmetry
% specify crystal and specimen symmetry CS = {... 'Not Indexed',... crystalSymmetry('m-3m','mineral','Fe'),... % crystal symmetry phase 1 crystalSymmetry('m-3m','mineral','Mg')}; % crystal symmetry phase 2
import ebsd data
% file name fname = fullfile(mtexDataPath,'EBSD','85_829grad_07_09_06.txt'); ebsd = EBSD.load(fname,'CS',CS,... 'ColumnNames', { 'Phase' 'x' 'y' 'Euler 1' 'Euler 2' 'Euler 3' 'Mad' 'BC'},... 'Columns', [2 3 4 5 6 7 8 9],... 'ignorePhase', 0, 'Bunge');
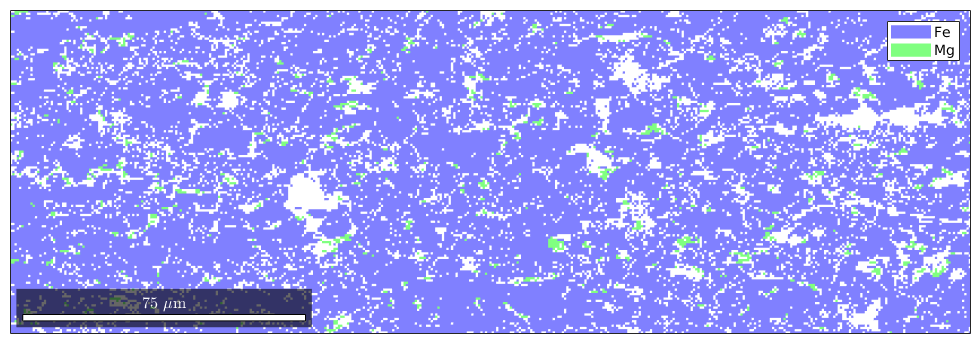
Plot Spatial Data
plot(ebsd)
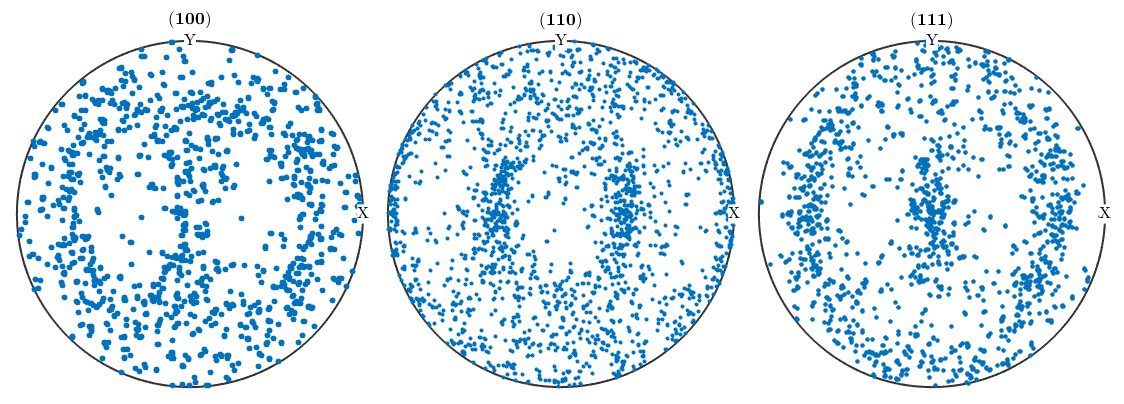
Plot Pole Figures as Scatter Plots
h = [Miller(1,0,0,CS{2}),Miller(1,1,0,CS{2}),Miller(1,1,1,CS{2})];
plotPDF(ebsd('Fe').orientations,h,'points',500,'antipodal')I'm plotting 500 random orientations out of 48184 given orientations You can specify the the number points by the option "points". The option "all" ensures that all data are plotted
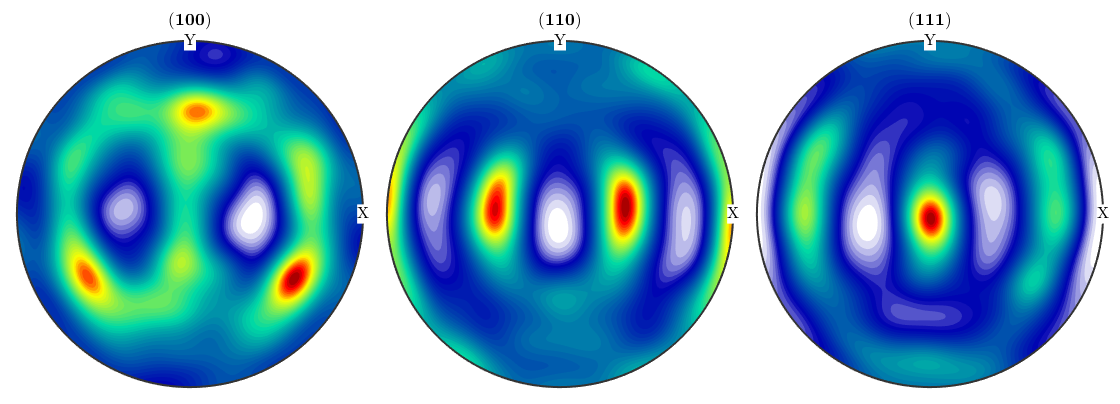
Kernel Density Estimation
The crucial point in kernel density estimation in the choice of the halfwidth of the kernel function used for estimation. If the halfwidth of is chosen to small the single orientations are visible rather then the ODF (compare plot of ODF1). If the halfwidth is chosen to wide the estimated ODF becomes very smooth (ODF2).
odf1 = calcODF(ebsd('Fe').orientations) odf2 = calcODF(ebsd('Fe').orientations,'halfwidth',5*degree)
odf1 = ODF
crystal symmetry : Fe (m-3m)
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
odf2 = ODF
crystal symmetry : Fe (m-3m)
specimen symmetry: 1
Harmonic portion:
degree: 48
weight: 1
Plot pole figures
plotPDF(odf1,h,'antipodal') figure plotPDF(odf2,h,'antipodal')

Plot ODF
plot(odf2,'sections',9,'resolution',2*degree,... 'FontSize',10,'silent')
Warning: Plot properties not compatible to previous plot! I'going to create a new figure.

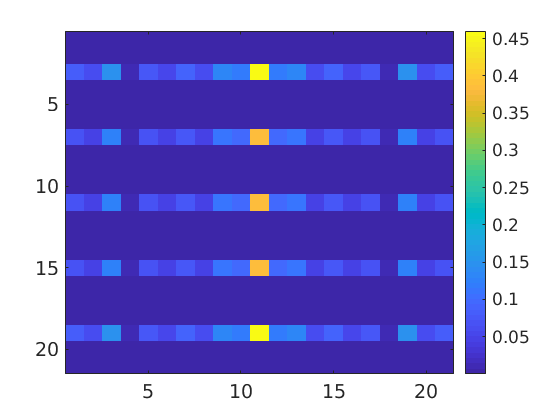
Estimation of Fourier Coefficients
Once, a ODF has been estimated from EBSD data it is straight forward to calculate Fourier coefficients. E.g. by
close all odf2 = FourierODF(odf2); imagesc(abs(odf2.calcFourier('order',10))) mtexColorbar
However this is a biased estimator of the Fourier coefficents which underestimates the true Fourier coefficients by a factor that correspondes to the decay rate of the Fourier coeffients of the kernel used for ODF estimation. One obtains an unbiased estimator of the Fourier coefficients if they are calculated from the ODF estimated with the help fo the Direchlet kernel. I.e.
dirichlet = DirichletKernel(32); odf3 = calcODF(ebsd('Fe').orientations,'kernel',dirichlet); imagesc(abs(odf3.calcFourier('order',10))) mtexColorbar
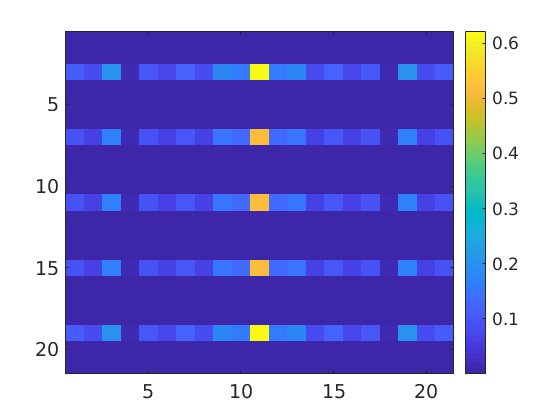
Let us compare the Fourier coefficients obtained by both methods.
plotFourier(odf2,'bandwidth',32) hold all plotFourier(odf3,'bandwidth',32) hold off

A Sythetic Example
Simulate EBSD data from a given standard ODF
CS = crystalSymmetry('trigonal'); fibre_odf = 0.5*uniformODF(CS) + 0.5*fibreODF(Miller(0,0,0,1,CS),zvector); plot(fibre_odf,'sections',6,'silent') ori = calcOrientations(fibre_odf,10000)
Warning: Plot properties not compatible to previous plot! I'going to create a new figure. ori = orientation size: 10000 x 1 crystal symmetry : -31m, X||a*, Y||b, Z||c* specimen symmetry: 1
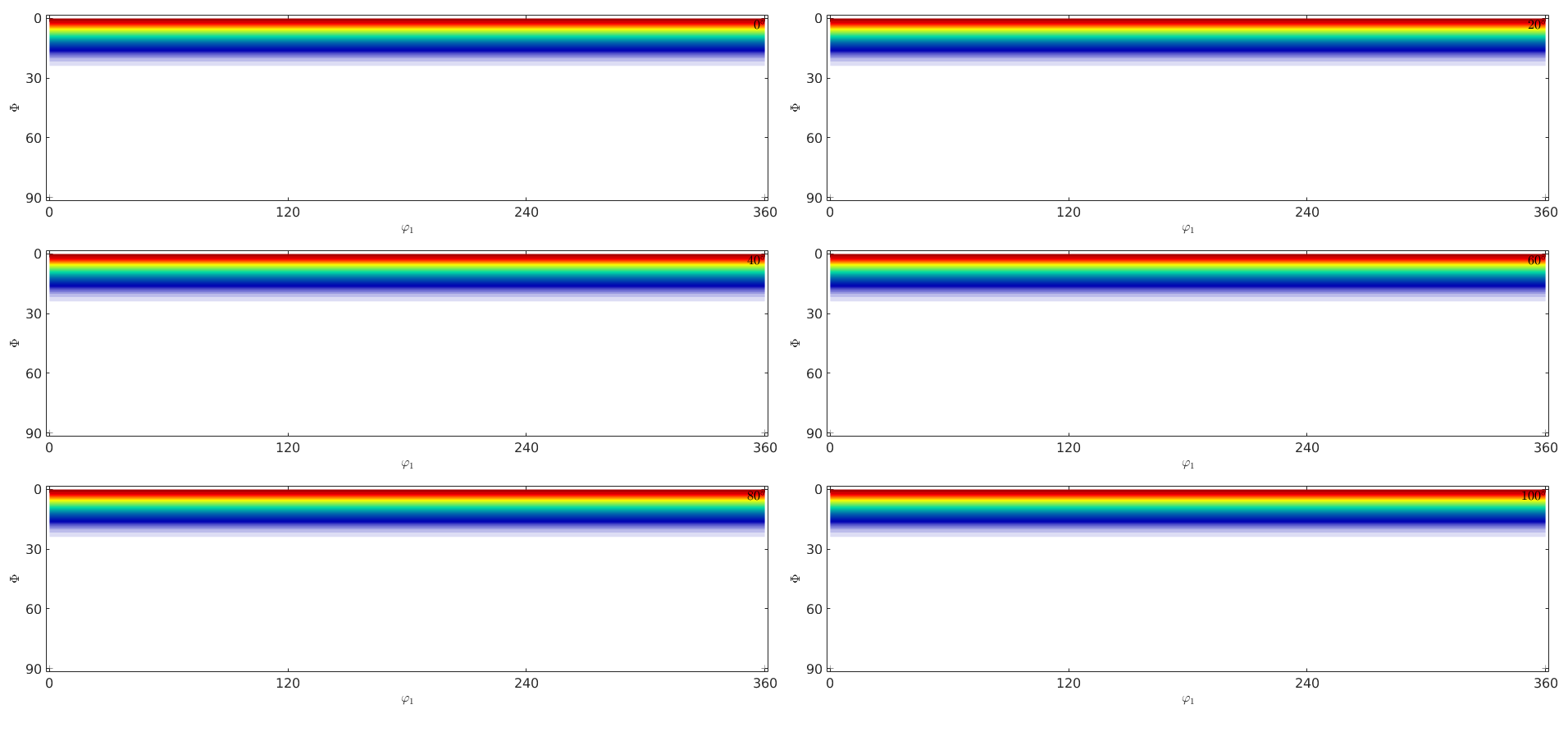
Estimate an ODF from the simulated EBSD data
odf = calcODF(ori)
odf = ODF
crystal symmetry : -31m, X||a*, Y||b, Z||c*
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
plot the estimated ODF
plot(odf,'sections',6,'silent')
Warning: Plot properties not compatible to previous plot! I'going to create a new figure.
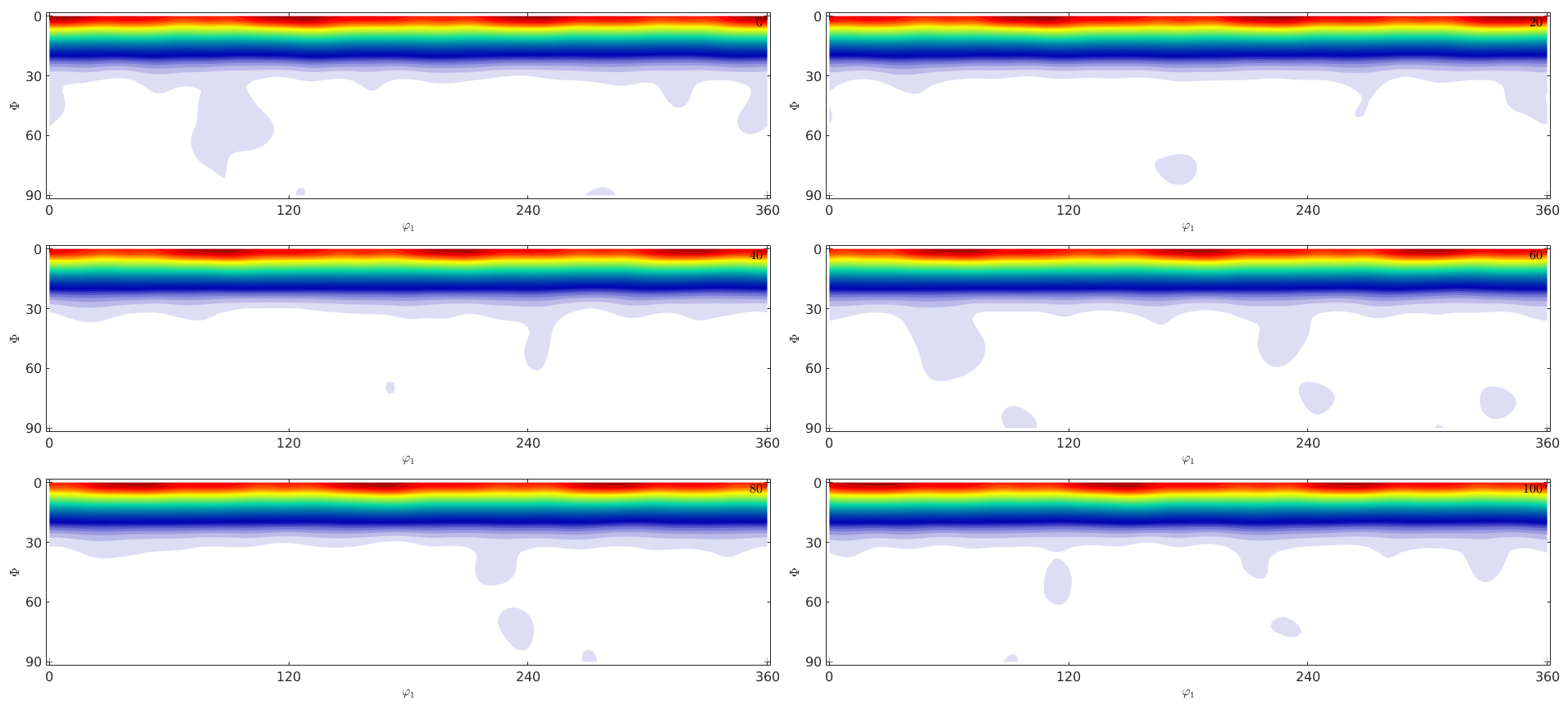
calculate estimation error
calcError(odf,fibre_odf,'resolution',5*degree)ans =
0.0945
For a more exhausive example see the EBSD Simulation demo!
Exercises
5)
a) Load the EBSD data: data/ebsd\_txt/85\_829grad\_07\_09\_06.txt!
import_wizard('ebsd')
b) Estimate an ODF from the above EBSD data.
odf = calcODF(ori)
odf = ODF
crystal symmetry : -31m, X||a*, Y||b, Z||c*
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
c) Visualize the ODF and some of its pole figures!
plot(odf,'sections',6,'silent')
Warning: Plot properties not compatible to previous plot! I'going to create a new figure.
d) Explore the influence of the halfwidth to the kernel density estimation by looking at the pole figures!
| DocHelp 0.1 beta |