Orientation Density Functions
This example demonstrates the most important MTEX tools for analysing ODFs.
All described commands can be applied to model ODFs constructed via uniformODF, unimodalODF, or fibreODF and to all estimated ODF calculated from pole figures or EBSD data.
Defining Model ODFs
Unimodal ODFs
SS = specimenSymmetry('orthorhombic'); CS = crystalSymmetry('cubic'); o = orientation.brass(CS,SS); psi = vonMisesFisherKernel('halfwidth',20*degree); odf1 = unimodalODF(o,CS,SS,psi)
odf1 = ODF
crystal symmetry : m-3m
specimen symmetry: mmm
Radially symmetric portion:
kernel: van Mises Fisher, halfwidth 20°
center: (35°,45°,0°)
weight: 1
Fibre ODFs
CS = crystalSymmetry('hexagonal'); h = Miller(1,0,0,CS); r = xvector; psi = AbelPoissonKernel('halfwidth',18*degree); odf2 = fibreODF(h,r,psi)
odf2 = ODF
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Fibre symmetric portion:
kernel: Abel Poisson, halfwidth 18°
fibre: (10-10) - 1,0,0
weight: 1
uniform ODFs
odf3 = uniformODF(CS)
odf3 = ODF
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Uniform portion:
weight: 1
FourierODF
Bingham ODFs
Lambda = [-10,-10,10,10] A = quaternion(eye(4)) odf = BinghamODF(Lambda,A,CS) plotIPDF(odf,xvector) plotPDF(odf,Miller(1,0,0,CS))
Lambda =
-10 -10 10 10
A = Quaternion
size: 1 x 4
a b c d
1 0 0 0
0 1 0 0
0 0 1 0
0 0 0 1
odf = ODF
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Bingham portion:
kappa: -10 -10 10 10
weight: 1
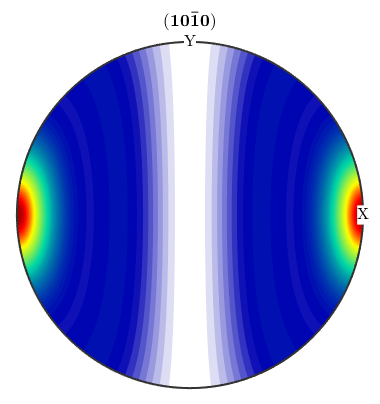
ODF Arithmetics
0.3*odf2 + 0.7*odf3 rot = rotation.byAxisAngle(yvector,90*degree); odf = rotate(odf,rot) plotPDF(odf,Miller(1,0,0,CS))
ans = ODF
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Fibre symmetric portion:
kernel: Abel Poisson, halfwidth 18°
fibre: (10-10) - 1,0,0
weight: 0.3
Uniform portion:
weight: 0.7
odf = ODF
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Bingham portion:
kappa: -10 -10 10 10
weight: 1
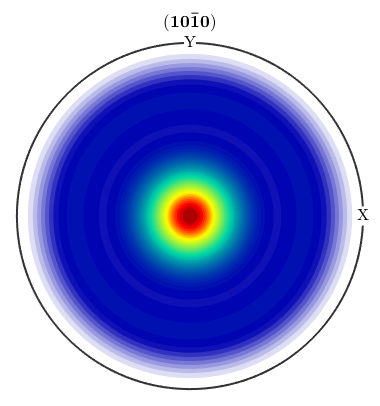
Working with ODFs
% *Texture Characteristics+ calcError(odf2,odf3,'L1') % difference between ODFs [maxODF,centerODF] = max(odf) % the modal orientation mean(odf) % the mean orientation max(odf) volume(odf,centerODF,5*degree) % the volume of a ball fibreVolume(odf2,h,r,5*degree) % the volume of a fibre textureindex(odf) % the texture index entropy(odf) % the entropy f_hat = calcFourier(odf2,16); % the C-coefficients up to order 16
ans =
0.2030
maxODF =
3.3783
centerODF = orientation
size: 1 x 1
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Bunge Euler angles in degree
phi1 Phi phi2 Inv.
90 90 270 0
ans = orientation
size: 1 x 1
crystal symmetry : 6/mmm, X||a*, Y||b, Z||c*
specimen symmetry: 1
Bunge Euler angles in degree
phi1 Phi phi2 Inv.
106.893 0.000738512 251.926 0
ans =
3.3783
ans =
0.0028
ans =
0.0253
ans =
1.9251
ans =
-0.5028
Plotting ODFs
Plotting (Inverse) Pole Figures
close all plotPDF(odf,Miller(0,1,0,CS),'antipodal') plotIPDF(odf,[xvector,zvector])
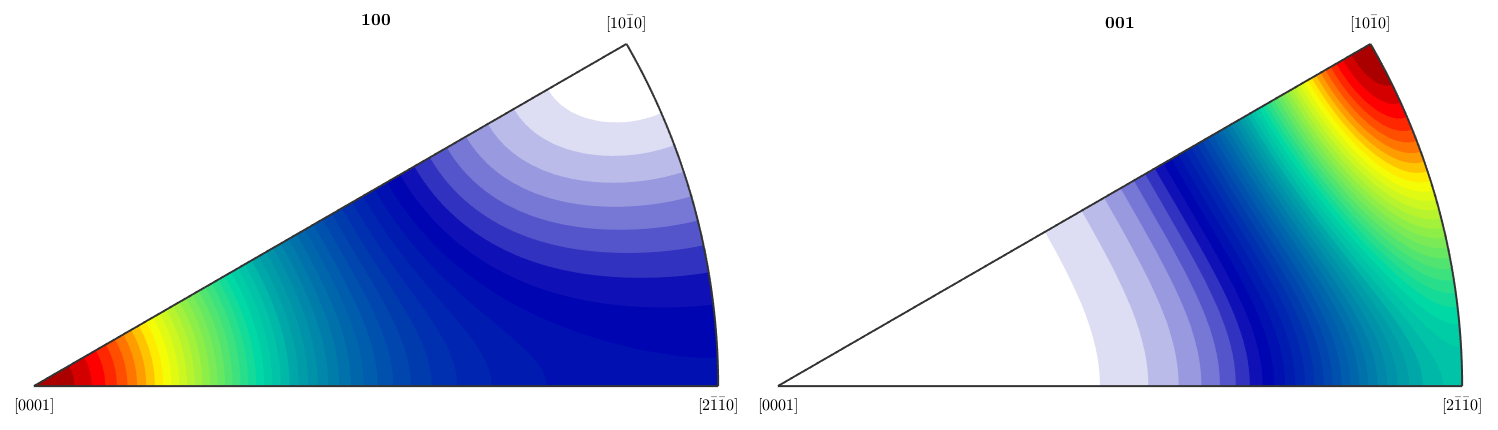
Plotting an ODF
close all plot(SantaFe,'alpha','sections',6,'projection','plain','contourf') mtexColorMap white2black
Exercises
2)
a) Construct a cubic unimodal ODF with mod at [0 0 1](3 1 0). (Miller indice). What is its modal orientation in Euler angles?
CS = crystalSymmetry('cubic');
ori = orientation.byMiller([0 0 1],[3 1 0],CS);
odf = unimodalODF(ori);b) Plot some pole figures. Are there pole figures with and without antipodal symmetry? What about the inverse pole figures?
plotPDF(odf,[Miller(1,0,0,CS),Miller(2,3,1,CS)])
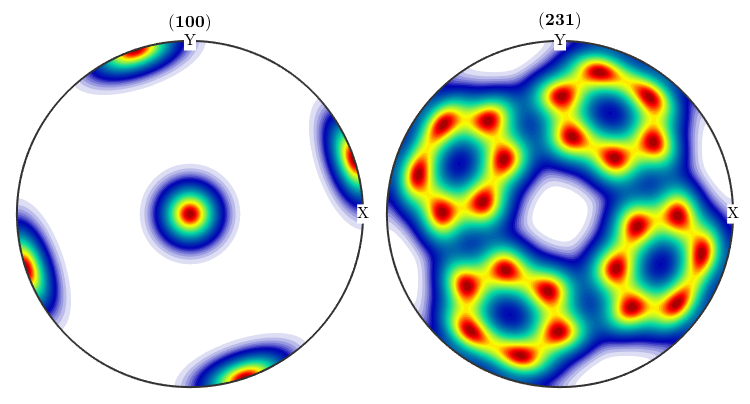
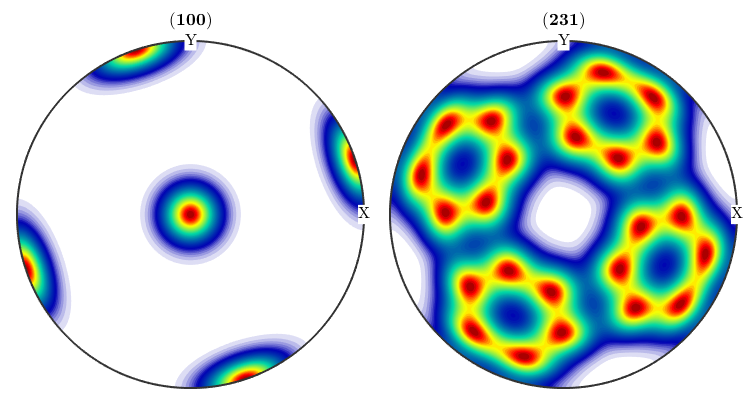
close all;plotPDF(odf,[Miller(1,0,0,CS),Miller(2,3,1,CS)],'antipodal')
close all;plotIPDF(odf,vector3d(1,1,3))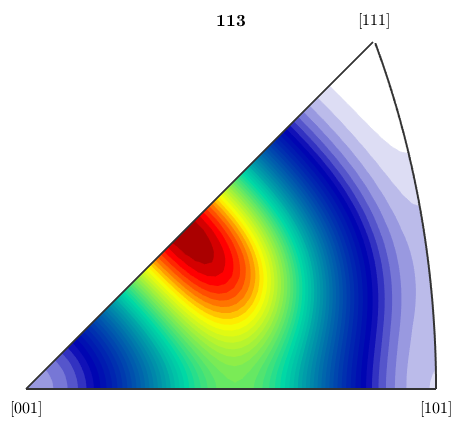
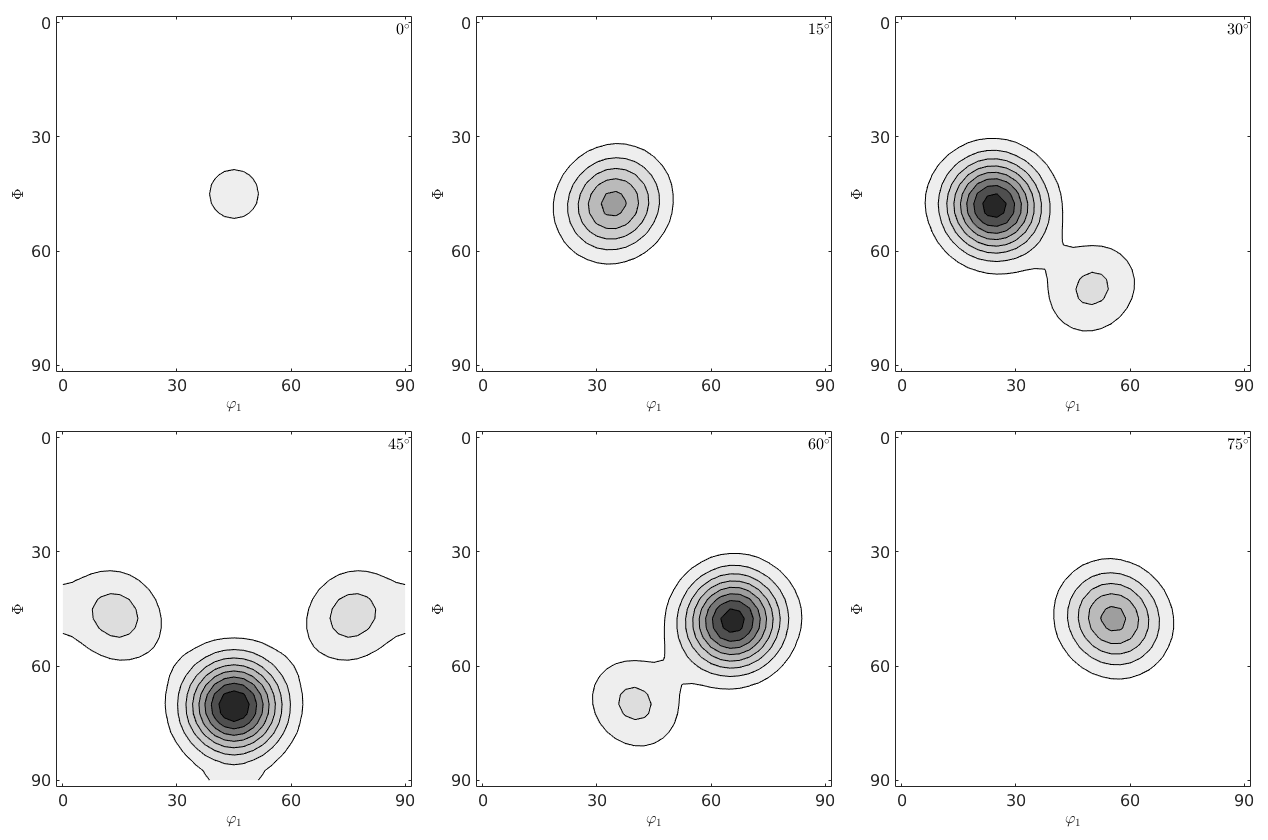
c) Plot the ODF in sigma and phi2 - sections. How many mods do you observe?
close all; plot(odf,'sections',6)
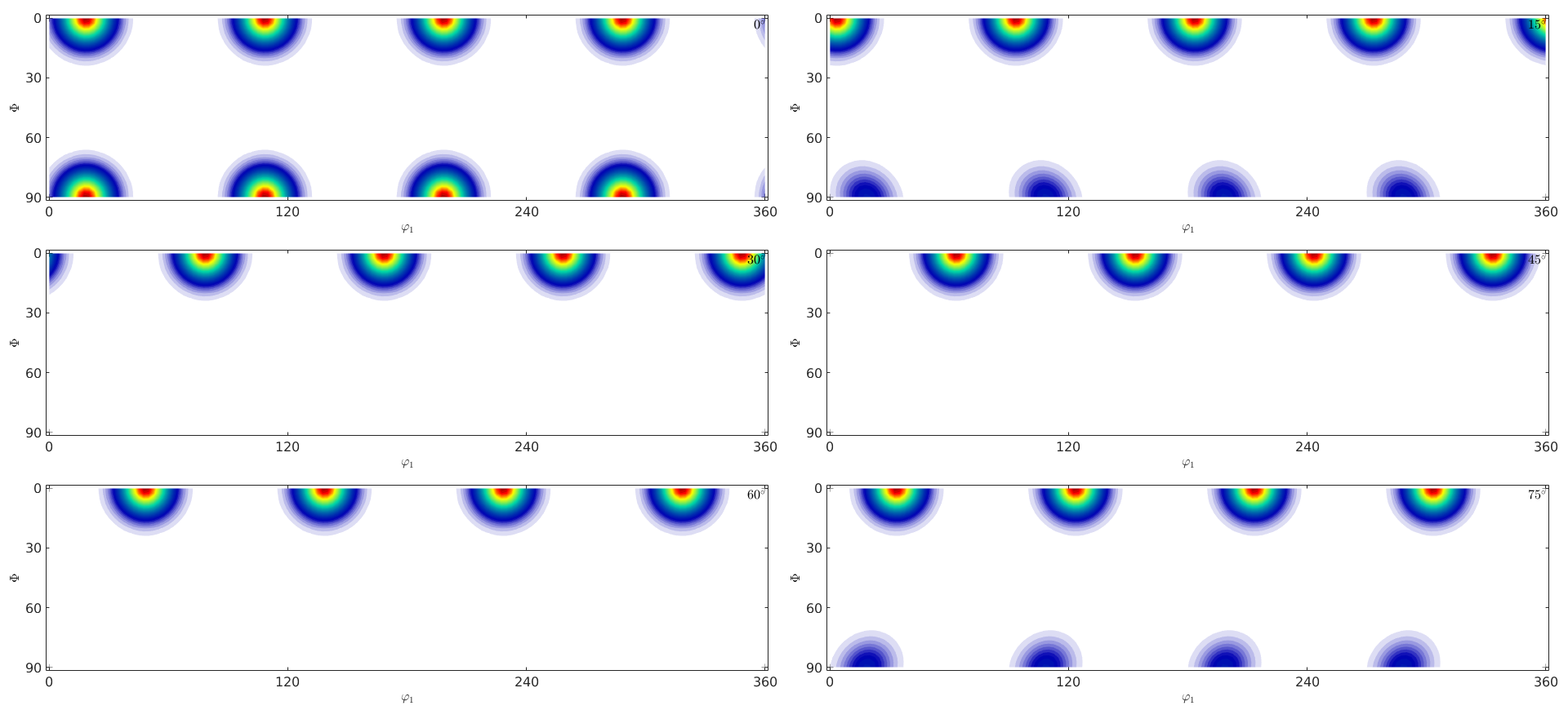
d) Compute the volume of the ODF that is within a distance of 10 degree of the mod. Compare to an the uniform ODF.
volume(odf,ori,10*degree) volume(uniformODF(CS,SS),ori,10*degree)
ans =
0.2939
ans =
0.0269
e) Construct a trigonal ODF that consists of two fibres at h1 = (0,0,0,1), r1 = (0,1,0), h2 = (1,0,-1,0), r2 = (1,0,0). Do the two fibres intersect?
cs = crystalSymmetry('trigonal'); odf = 0.5 * fibreODF(Miller(0,0,0,1,cs),yvector) + ... 0.5 * fibreODF(Miller(1,0,-1,0,cs),xvector)
odf = ODF
crystal symmetry : -31m, X||a*, Y||b, Z||c*
specimen symmetry: 1
Fibre symmetric portion:
kernel: de la Vallee Poussin, halfwidth 10°
fibre: (0001) - 0,1,0
weight: 0.5
Fibre symmetric portion:
kernel: de la Vallee Poussin, halfwidth 10°
fibre: (10-10) - 1,0,0
weight: 0.5
f) What is the modal orientation of the ODF?
mod = calcModes(odf)
mod = orientation size: 1 x 1 crystal symmetry : -31m, X||a*, Y||b, Z||c* specimen symmetry: 1 Bunge Euler angles in degree phi1 Phi phi2 Inv. 180 90 180 0
g) Plot the ODF in sigma and phi2 - sections. How many fibre do you observe?
close all;plot(odf,'sections',6) mtexColorMap white2black annotate(mod,'MarkerColor','r','Marker','s')
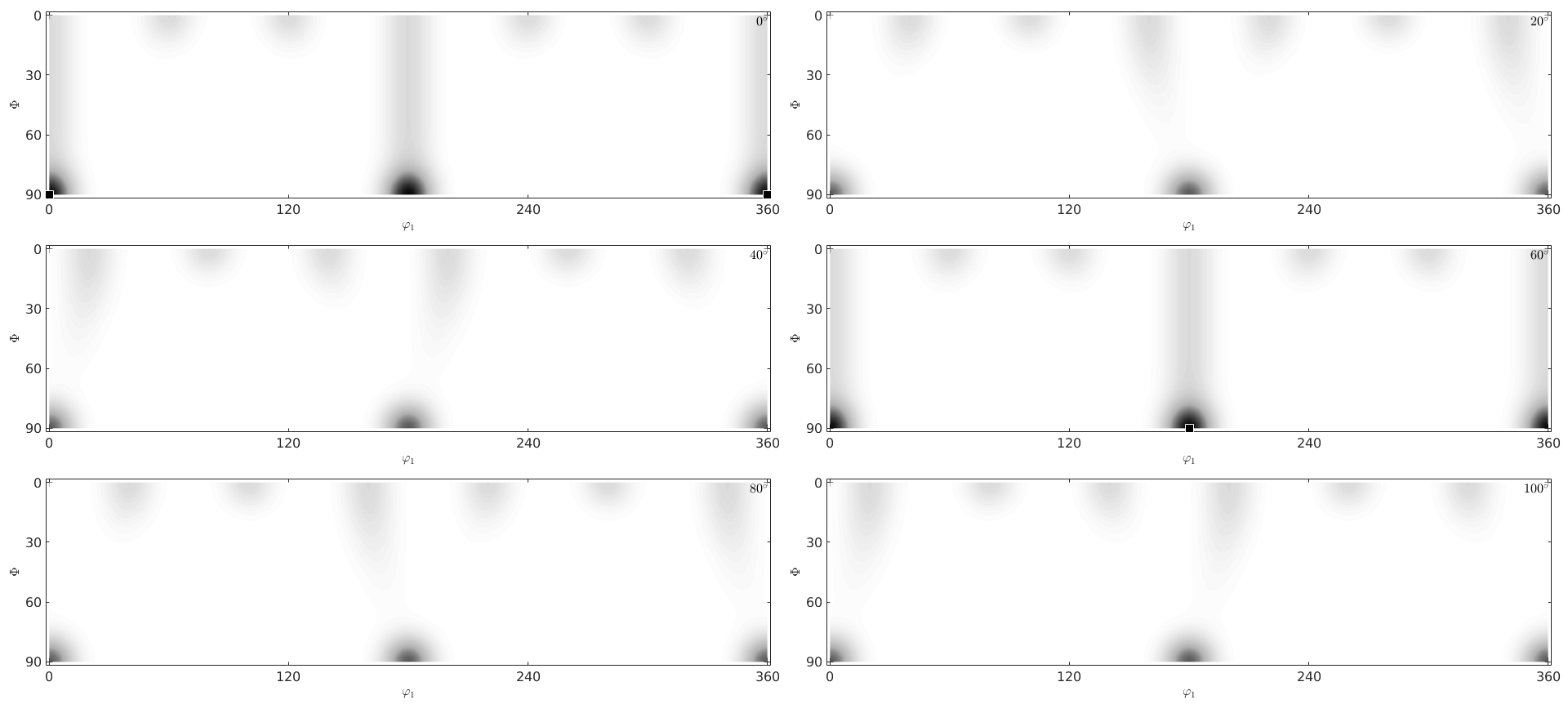
plot(odf,'phi2','sections',6)

h) Compute the texture index of the ODF.
textureindex(odf)
ans =
9.6324
| DocHelp 0.1 beta |