Fundamental Regions
Thanks to crystal symmetry the orientation space can be reduced to the so called fundamental or asymmetric region. Those regions play an important role for the computation of axis and angle distributions of misorientations.
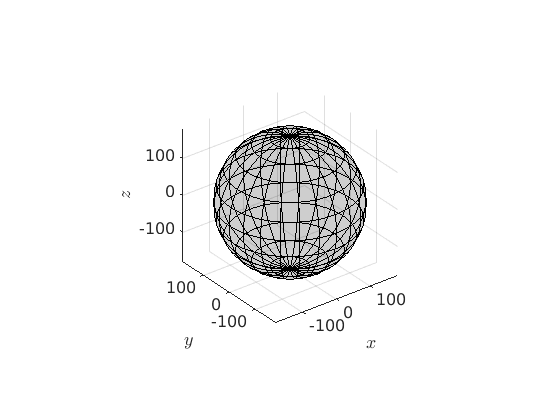
The complete, symmetry free, orientation space
The space orientations can be imagined as a three dimensional ball with radius 180 degree. The distance of some point in the ball to the origin represent the rotational angle and the vector represents the rotational axis. In MTEX this can be represented as follows
% triclic crystal symmetry cs = crystalSymmetry('triclinic') % the corresponding orientation space oR_all = fundamentalRegion(cs); % lets plot the ball of all orientations plot(oR_all)
cs = crystalSymmetry symmetry : -1 a, b, c : 1, 1, 1 alpha, beta, gamma: 90°, 90°, 90° reference frame : X||a, Y||b*, Z||c*
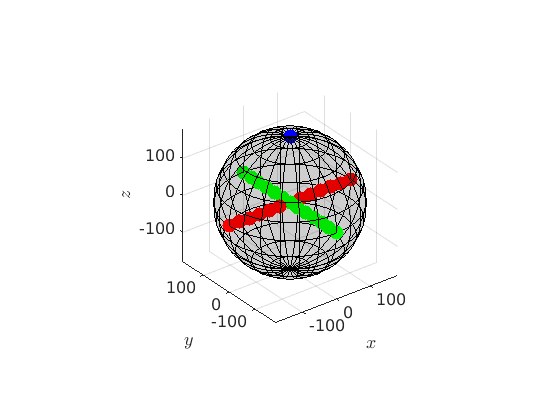
Next we plot some orientations into this space
% rotation about the z-axis about 180 degree rotZ = orientation.byAxisAngle(zvector,180*degree,cs); hold on plot(rotZ,'MarkerColor','b','MarkerSize',10) hold off % rotations about the x- and y-axis about 30,60,90 ... degree rotX = orientation.byAxisAngle(xvector,(-180:30:180)*degree,cs); rotY = orientation.byAxisAngle(yvector,(-180:30:180)*degree,cs); hold on plot(rotX,'MarkerColor','r','MarkerSize',10) plot(rotY,'MarkerColor','g','MarkerSize',10) hold off
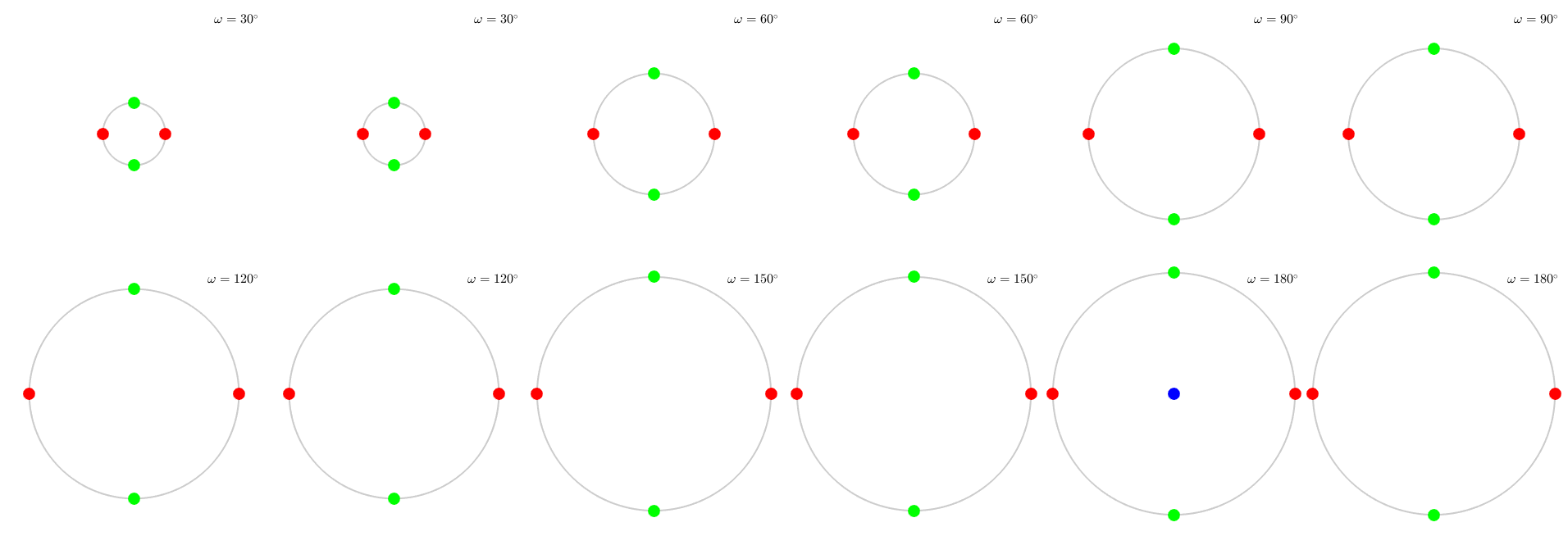
An alternative view on the orientation space is by sections, e.g. sections along the Euler angles or along the rotational angle. The latter one should be demonstrated next:
plotSection(rotZ,'MarkerColor','b','axisAngle',(30:30:180)*degree) hold on plot(rotX,'MarkerColor','g','add2all') hold on plot(rotY,'MarkerColor','r','add2all') hold off
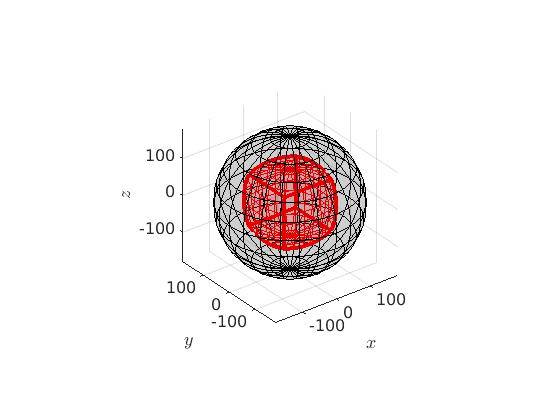
Crystal Symmetries
In case of crystal symmetries the orientation space can divided into as many equivalent segments as the symmetry group has elements. E.g. in the case of orthorombic symmetry the orientation space is subdivided into four equal parts, the central one looking like
cs = crystalSymmetry('222') oR = fundamentalRegion(cs); close all plot(oR_all) hold on plot(oR,'color','r') hold off
cs = crystalSymmetry symmetry: 222 a, b, c : 1, 1, 1 Warning: Possible symmetry mismach!
As an example consider the following EBSD data set
mtexdata forsteritewe can visualize the Forsterite orientations by
plot(ebsd('Fo').orientations,'axisAngle')
plot 2000 random orientations out of 152345 given orientations
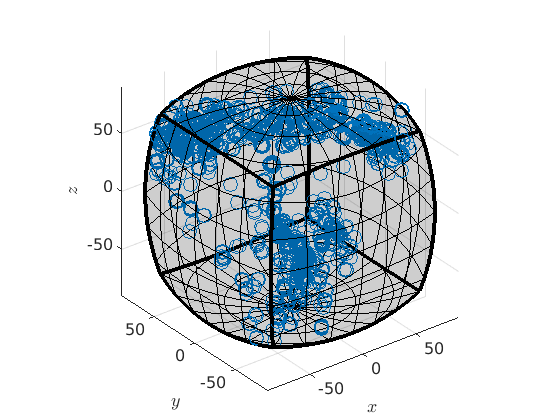
We see that all orientations are automatically projected inside the fundamental region. In order to compute explicitly the represent inside the fundamental region we can do
ori = ebsd('Fo').orientations.project2FundamentalRegionori = orientation size: 152345 x 1 crystal symmetry : Forsterite (mmm) specimen symmetry: 1
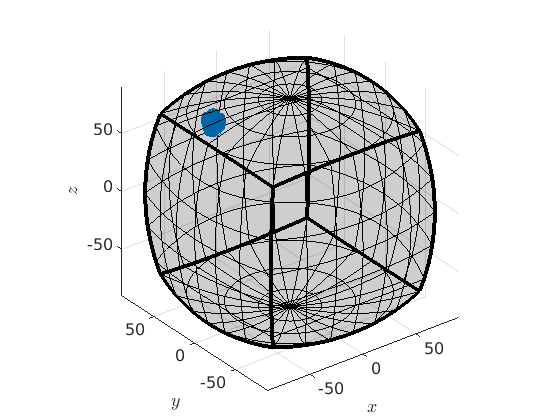
Change the center of the fundamental region
There is no necessity that the fundamental region has to be centered in the origin - it can be centered at any orientation, e.g. at the mean orientation of a grain.
% segment data into grains [grains,ebsd.grainId] = calcGrains(ebsd('indexed')); % take the orientations of the largest on [~,id] = max(grains.area); largeGrain = grains(id) ori = ebsd(largeGrain).orientations % recenter the fundamental zone to the mean orientation plot(rotate(oR,largeGrain.meanOrientation)) % project the orientations into the fundamental region around the mean % orientation ori = ori.project2FundamentalRegion(largeGrain.meanOrientation) plot(ori,'axisAngle')
largeGrain = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 1 2683 Forsterite mmm
boundary segments: 714
triple points: 44
Id Phase Pixels GOS phi1 Phi phi2
931 1 2683 0.0425468 171 55 262
ori = orientation
size: 2683 x 1
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
ori = orientation
size: 2683 x 1
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
plot 2000 random orientations out of 2683 given orientations
Fundamental regions of misorientations
Misorientations are characterised by two crystal symmetries. A corresponding fundamental region is defined by
oR = fundamentalRegion(ebsd('Fo').CS,ebsd('En').CS); plot(oR)
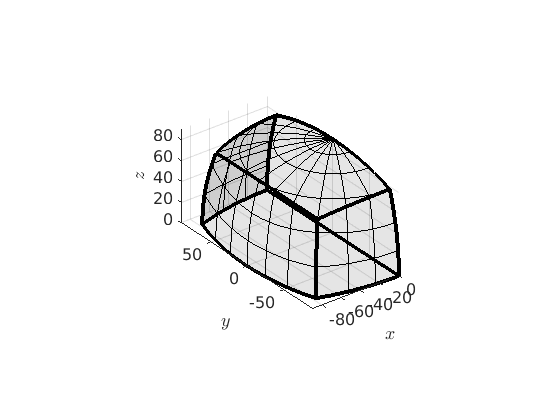
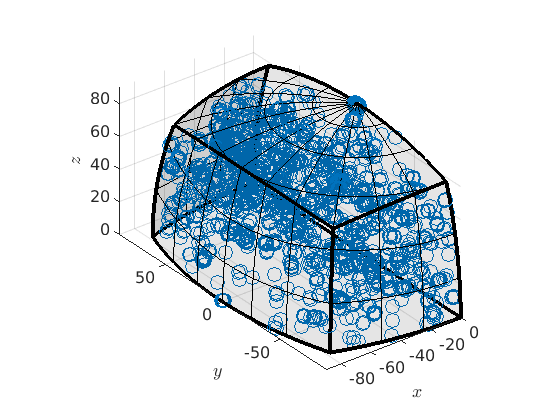
Let plot grain boundary misorientations within this fundamental region
plot(grains.boundary('fo','En').misorientation)
plot 2000 random orientations out of 11814 given orientations
Fundamental regions of misorientations with antipodal symmetry
Note that for boundary misorientations between the same phase we can not distinguish between a misorientation and its inverse. This is not the case for misorientations between different phases or the misorientation between the mean orientation of a grain and all other orientations. The inverse of a misorientation is axis - angle representation is simply the one with the same angle but antipodal axis. Accordingly this additional symmetry is handled in MTEX by the keyword antipodal.
oR = fundamentalRegion(ebsd('Fo').CS,ebsd('Fo').CS,'antipodal'); plot(oR)
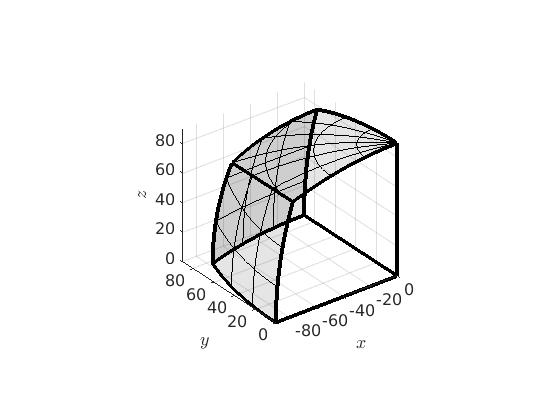
We see that the fundamental region with antipodal symmetry has only half the size as without. In the case of misorientations between the same phase MTEX automatically sets the antipodal flag to the misorientations and plots them accordingly.
mori = grains.boundary('Fo','Fo').misorientation plot(mori)
mori = misorientation size: 15974 x 1 crystal symmetry : Forsterite (mmm) crystal symmetry : Forsterite (mmm) antipodal: true plot 2000 random orientations out of 15974 given orientations
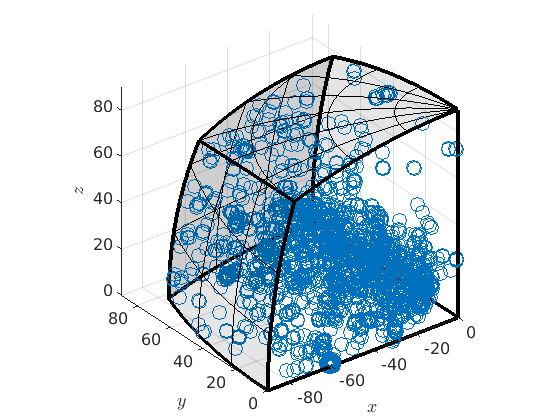
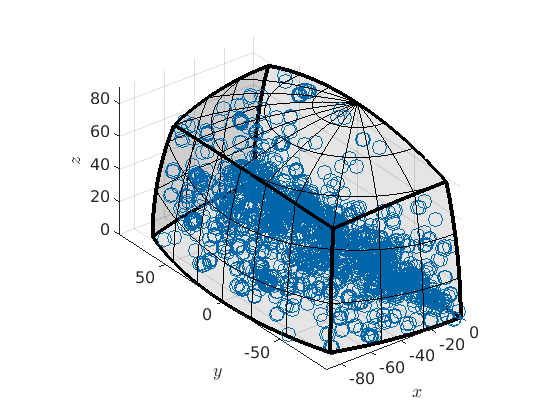
If you want to avoid this you can remove the anitpodal flag by
mori.antipodal = false; plot(mori)
plot 2000 random orientations out of 15974 given orientations
Axis angle sections
Again we can plot constant angle sections through the fundamental region. This is done by
plotSection(mori,'axisAngle')plotting 2000 random orientations out of 15974 given orientations
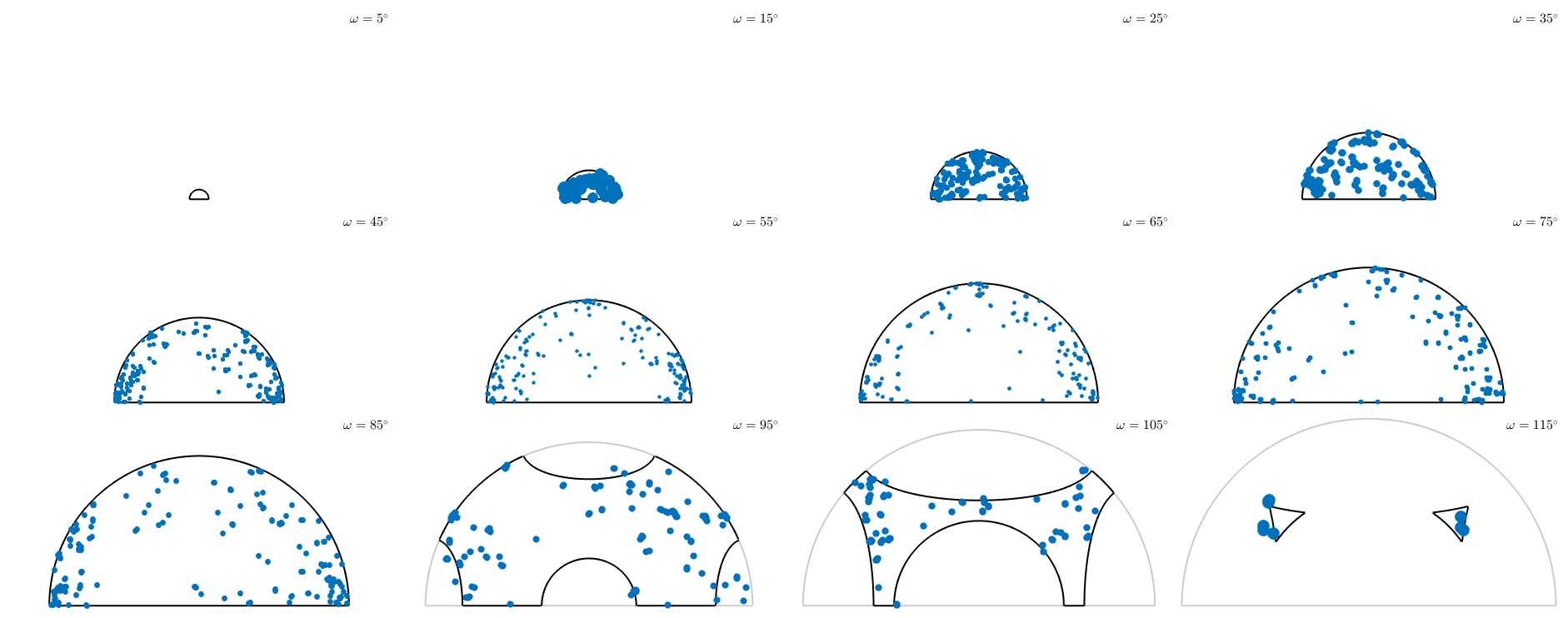
Note that in the previous plot we distinguish between mori and inv(mori). Adding antipodal symmetry those are considered as equivalent
plotSection(mori,'axisAngle','antipodal')
plotting 2000 random orientations out of 15974 given orientations
| DocHelp 0.1 beta |