Misorientation Distribution Function
Explains how to compute and analyze misorientation distribution functions.
Computing a misorientation distribution function from EBSD data
Lets import some EBSD data and reconstruct the grains.
mtexdata forsterite
grains = calcGrains(ebsd)
grains = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
0 16334 58485 notIndexed
1 4092 152345 Forsterite mmm
2 1864 26058 Enstatite mmm
3 1991 9064 Diopside 12/m1 X||a*, Y||b*, Z||c
boundary segments: 147957
triple points: 11456
Properties: GOS, meanRotation
The boundary misorientation distribution function
The boundary misorientation distribution function for the phase transition from Forsterite to Enstatite can be computed by
mdf_boundary = calcODF(grains.boundary('Fo','En').misorientation,'halfwidth',10*degree)
mdf_boundary = MDF
crystal symmetry : Forsterite (mmm)
crystal symmetry : Enstatite (mmm)
Harmonic portion:
degree: 25
weight: 1
The misorientation distribution function can be processed as any other ODF. E.g. we can compute the prefered misorientation via
[v,mori] = max(mdf_boundary)
v =
38.8766
mori = misorientation
size: 1 x 1
crystal symmetry : Forsterite (mmm)
crystal symmetry : Enstatite (mmm)
Bunge Euler angles in degree
phi1 Phi phi2 Inv.
82.4065 1.06577 187.671 0
or plot the pole figure corresponding to the crystal axis (1,0,0)
plotPDF(mdf_boundary,Miller(1,0,0,ebsd('Fo').CS))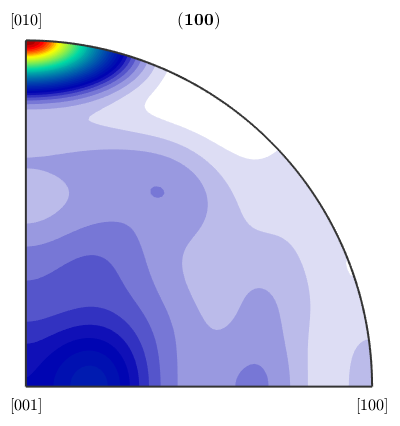
The uncorrelated misorientation distribution function
Alternatively the uncorrelated misorientation distribution function can be computed by providing the option uncorrelated
mori = calcMisorientation(ebsd('En'),ebsd('Fo')) mdf_uncor = calcODF(mori)
mori = misorientation
size: 98541 x 1
crystal symmetry : Forsterite (mmm)
crystal symmetry : Enstatite (mmm)
mdf_uncor = MDF
crystal symmetry : Forsterite (mmm)
crystal symmetry : Enstatite (mmm)
Harmonic portion:
degree: 25
weight: 1
Obviously it is different from the boundary misorientation distribution function.
plotPDF(mdf_uncor,Miller(1,0,0,ebsd('Fo').CS))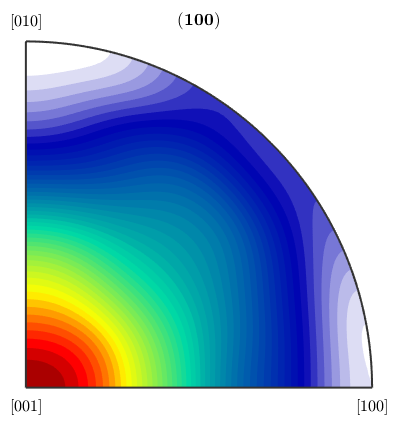
Computing the uncorrelated misorientation function from two ODFs
Let given two odfs
odf_fo = calcODF(ebsd('fo').orientations,'halfwidth',10*degree) odf_en = calcODF(ebsd('en').orientations,'halfwidth',10*degree)
odf_fo = ODF
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
odf_en = ODF
crystal symmetry : Enstatite (mmm)
specimen symmetry: 1
Harmonic portion:
degree: 25
weight: 1
Then the uncorrelated misorientation function between these two ODFs can be computed by
mdf = calcMDF(odf_en,odf_fo)
mdf = MDF
crystal symmetry : Forsterite (mmm)
crystal symmetry : Enstatite (mmm)
Harmonic portion:
degree: 19
weight: 1
This misorientation distribution function should be similar to the uncorrelated misorientation function computed directly from the ebsd data
plotPDF(mdf,Miller(1,0,0,ebsd('Fo').CS))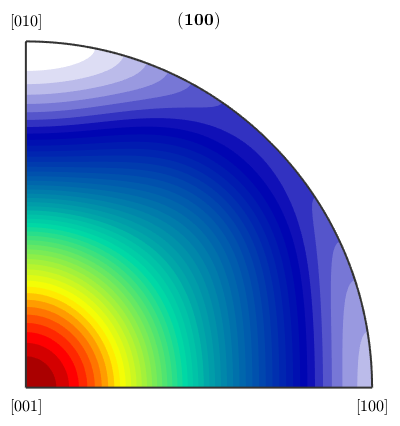
Analyzing misorientation functions
Angle distribution
Let us first compare the actual angle distribution of the boundary misorientations with the theoretical angle distribution of the uncorrelated MDF.
close all plotAngleDistribution(grains.boundary('fo','en').misorientation) hold on plotAngleDistribution(mdf) hold off
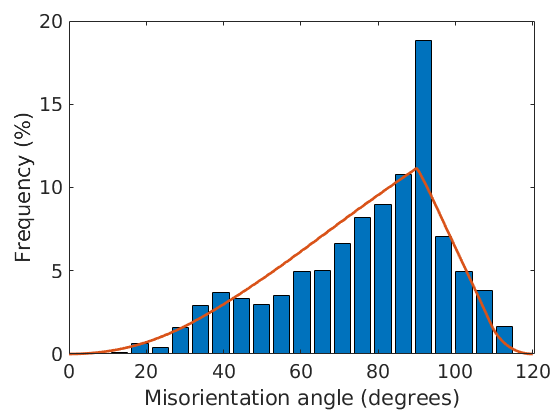
For computing the exact values see the commands calcAngleDistribution(mdf) and calcAngleDistribution(grains).
Axis distribution
The same we can do with the axis distribution. First the actual angle distribution of the boundary misorientations
plotAxisDistribution(grains.boundary('fo','en').misorientation,'smooth')
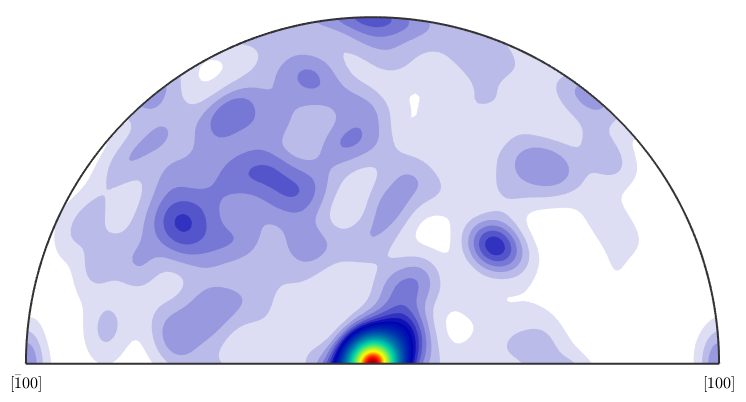
Now the theoretical axis distribution of the uncorrelated MDF.
plotAxisDistribution(mdf)
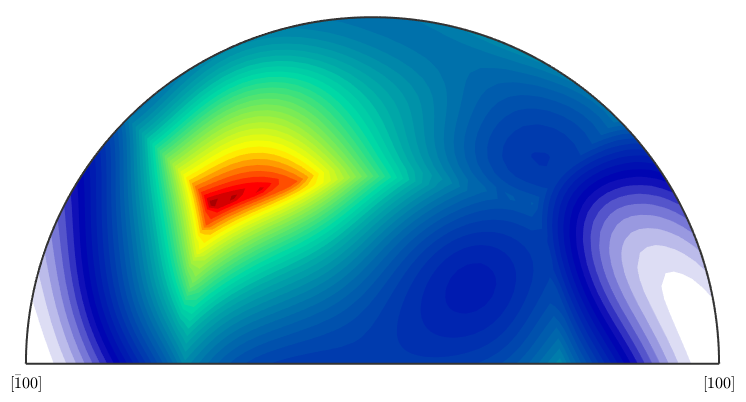
For computing the exact values see the commands calcAxisDistribution(mdf) and calcAxisDistribution(grains).
| DocHelp 0.1 beta |