Working with Grains
How to index grains and access shape properties.
| On this page ... |
| Accessing individual grains |
| Indexing by a Condition |
| Indexing by orientation or position |
| Grain-size Analysis |
| Spatial Dependencies |
Grains have several intrinsic properties, which can be used for statistical, shape as well as for spatial analysis
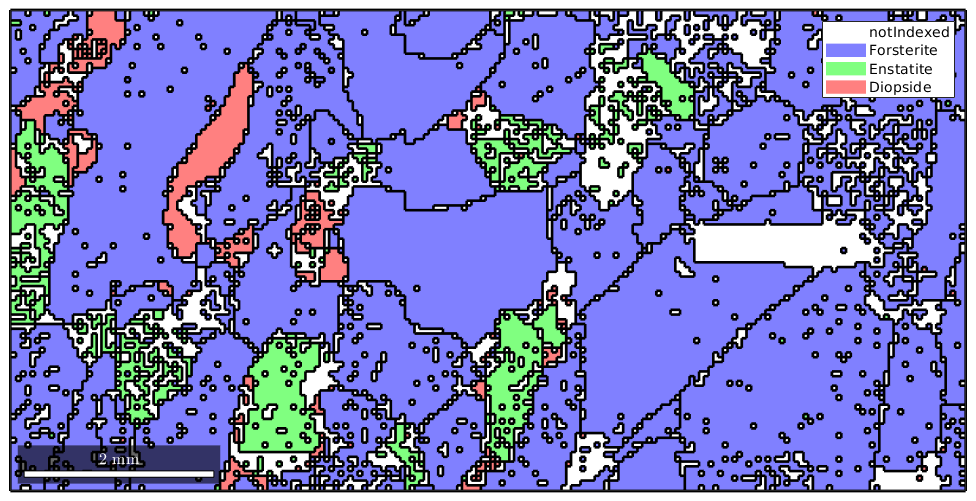
Let us first import some EBSD data and reconstruct grains
plotx2east mtexdata forsterite ebsd = ebsd(inpolygon(ebsd,[5 2 10 5]*10^3)); grains = calcGrains(ebsd) plot(ebsd) hold on plot(grains.boundary,'linewidth',2) hold off
grains = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
0 1139 4052 notIndexed
1 242 14093 Forsterite mmm
2 177 1397 Enstatite mmm
3 104 759 Diopside 12/m1 X||a*, Y||b*, Z||c
boundary segments: 10420
triple points: 903
Properties: GOS, meanRotation
Accessing individual grains
The variable grains is essentially a large vector of grains. Thus when applying a function like area to this variable we obtain a vector of the same lenght with numbers representing the area of each grain
grain_area = grains.area;
As a first rather simple application we could colorize the grains according to their area, i.e., according to the numbers stored in grain_area
plot(grains,grain_area)
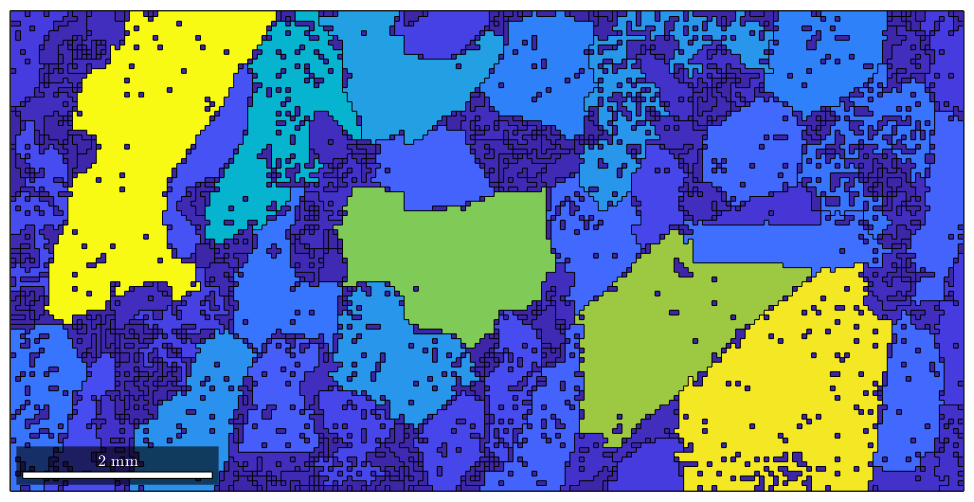
As a second application, we can ask for the largest grain within our data set. The maximum value and its position within a vector are found by the Matlab command max.
[max_area,max_id] = max(grain_area)
max_area =
3.8945e+06
max_id =
1617
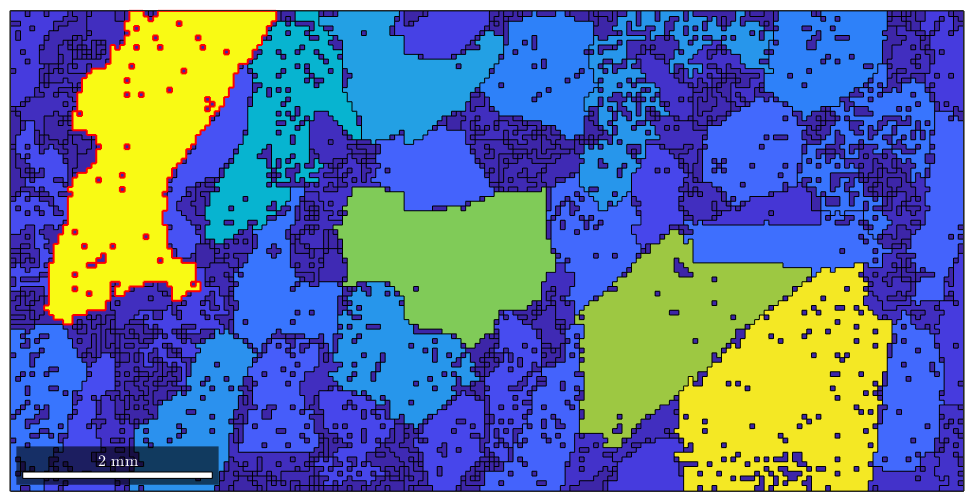
The number max_id is the position of the grain with a maximum area within the variable grains. We can access this specific grain by direct indexing
grains(max_id)
ans = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 1 1548 Forsterite mmm
boundary segments: 424
triple points: 70
Id Phase Pixels GOS phi1 Phi phi2
1617 1 1548 0.0129383 167 81 251
and so we can plot it
hold on plot(grains(max_id).boundary,'linecolor','red','linewidth',1.5) hold off
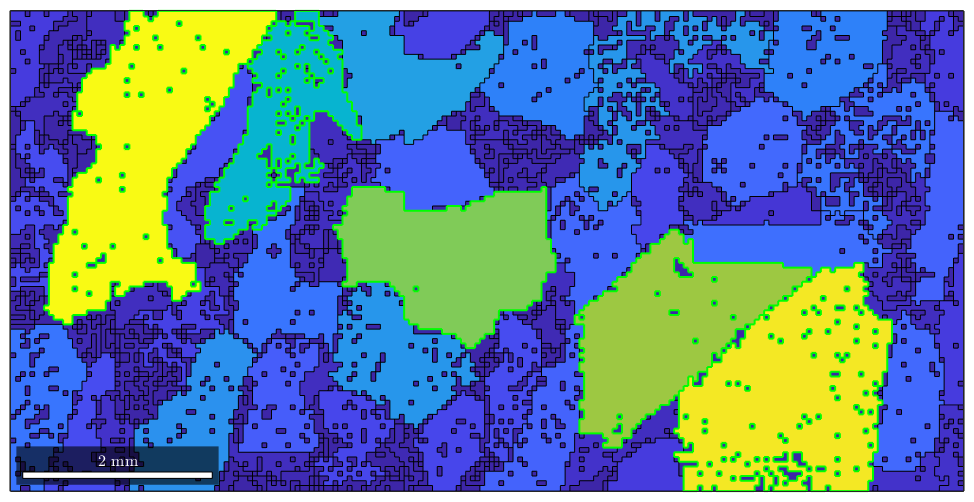
Note that this way of addressing individual grains can be generalized to many grains. E.g. assume we are interested in the largest 5 grains. Then we can sort the vector grain_area and take the indices of the 5 largest grains.
[sorted_area,sorted_id] = sort(grain_area,'descend'); large_grain_id = sorted_id(1:5); hold on plot(grains(large_grain_id).boundary,'linecolor','green','linewidth',1.5) hold off
Indexing by a Condition
By the same syntax as above we can also single out grains that satisfy a certain condition. I.e., to access are grains that are at least half as large as the largest grain we can do
condition = grain_area > max_area/2; hold on plot(grains(condition).boundary,'linecolor','red','linewidth',1.5) hold off
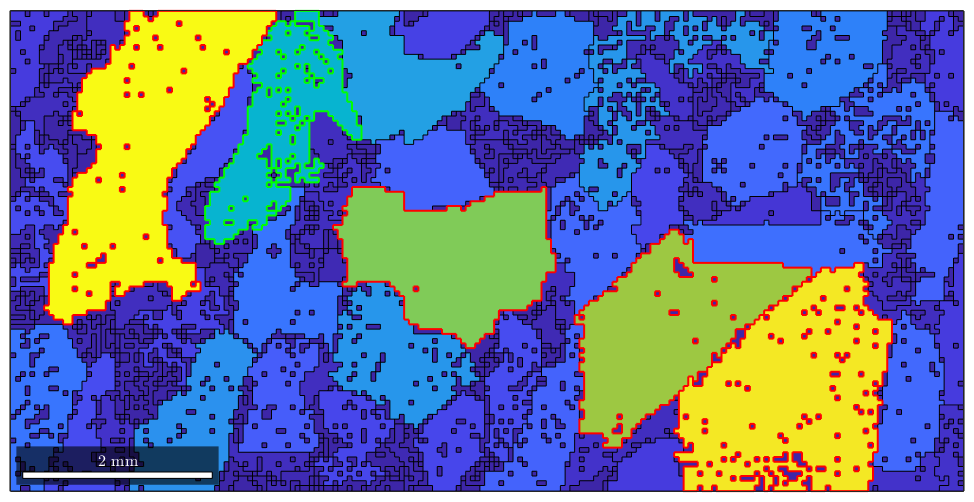
This is a very powerful way of accessing grains as the condition can be build up using any grain property. As an example let us consider the phase. The phase of the first five grains we get by
grains(1:5).phase
ans =
0
0
0
0
2
Now we can access or grains of the first phase Forsterite by the condition
condition = grains.phase == 1; plot(grains(condition))
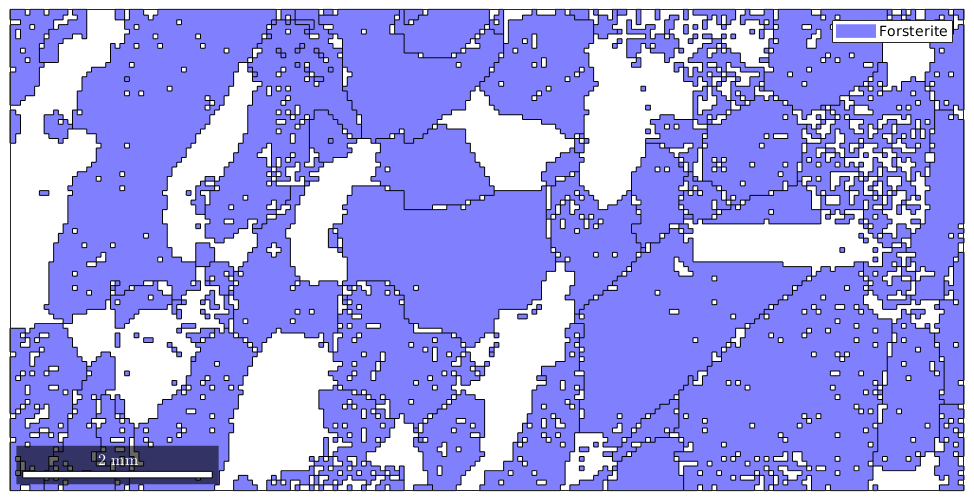
To make the above more directly you can use the mineral name for indexing
grains('forsterite')
ans = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 242 14093 Forsterite mmm
boundary segments: 7670
triple points: 795
Properties: GOS, meanRotation
Logical indexing allows also for more complex queries, e.g. selecting all grains perimeter larger than 6000 and at least 600 measurements within
condition = grains.perimeter>6000 & grains.grainSize >= 600; selected_grains = grains(condition) plot(selected_grains)
selected_grains = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 5 5784 Forsterite mmm
boundary segments: 1893
triple points: 244
Id Phase Pixels GOS phi1 Phi phi2
798 1 1447 0.013232 166 127 259
876 1 1075 0.00805971 153 68 237
999 1 1044 0.00753116 89 99 224
1617 1 1548 0.0129383 167 81 251
1620 1 670 0.0870135 143 78 274
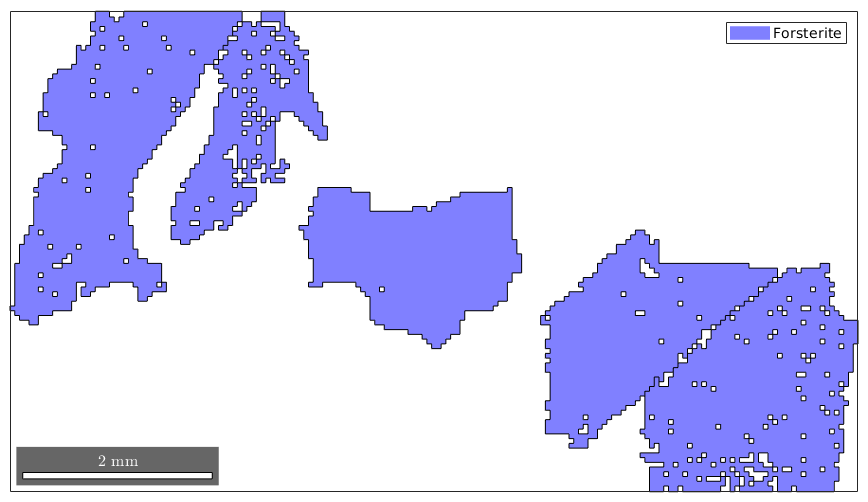
Indexing by orientation or position
One can also select a grain by its spatial coordinates using the syntax grains(x,y)
x = 12000; y = 4000; plot(grains); hold on plot(grains(x,y).boundary,'linewidth',2,'linecolor','r') plot(x,y,'marker','s','markerfacecolor','k',... 'markersize',10,'markeredgecolor','w') hold off
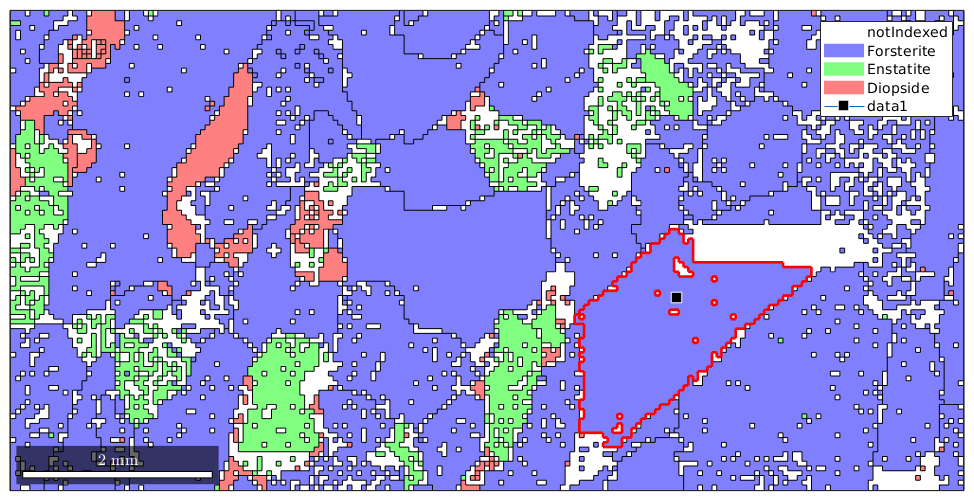
In order to select all grains with a certain orientation one can do
% restrict first to Forsterite phase grains_fo = grains('fo') % the reference orientation ori = orientation.byEuler(350*degree,50*degree,100*degree,grains('fo').CS) % select all grain with misorientation angle to ori less then 20 degree grains_selected = grains_fo(angle(grains_fo.meanOrientation,ori)<20*degree) plot(grains_selected)
grains_fo = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 242 14093 Forsterite mmm
boundary segments: 7670
triple points: 795
Properties: GOS, meanRotation
ori = orientation
size: 1 x 1
crystal symmetry : Forsterite (mmm)
specimen symmetry: 1
Bunge Euler angles in degree
phi1 Phi phi2 Inv.
350 50 100 0
grains_selected = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 12 2979 Forsterite mmm
boundary segments: 1603
triple points: 173
Id Phase Pixels GOS phi1 Phi phi2
359 1 63 0.0129176 177 140 250
389 1 1 0 167 128 260
622 1 352 0.00896432 153 122 252
636 1 305 0.0156453 164 115 268
680 1 331 0.0198713 152 143 247
798 1 1447 0.013232 166 127 259
1297 1 289 0.0315393 166 147 260
1549 1 1 0 1 47 279
1609 1 48 0.0169152 158 139 259
1629 1 10 0.0193004 172 125 261
1660 1 129 0.00814837 1 47 279
1662 1 3 0.00475795 1 47 279
Grain-size Analysis
Let's go back to the grain size and analyze its distribution. To this end, we consider the complete data set.
mtexdata forsterite % consider only indexed data for grain segmentation ebsd = ebsd('indexed'); % perform grain segmentation [grains,ebsd.grainId] = calcGrains(ebsd)
grains = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 1080 152345 Forsterite mmm
2 515 26058 Enstatite mmm
3 1496 9064 Diopside 12/m1 X||a*, Y||b*, Z||c
boundary segments: 43912
triple points: 3417
Properties: GOS, meanRotation
ebsd = EBSD
Phase Orientations Mineral Color Symmetry Crystal reference frame
1 152345 (81%) Forsterite light blue mmm
2 26058 (14%) Enstatite light green mmm
3 9064 (4.8%) Diopside light red 12/m1 X||a*, Y||b*, Z||c
Properties: bands, bc, bs, error, mad, x, y, grainId
Scan unit : um
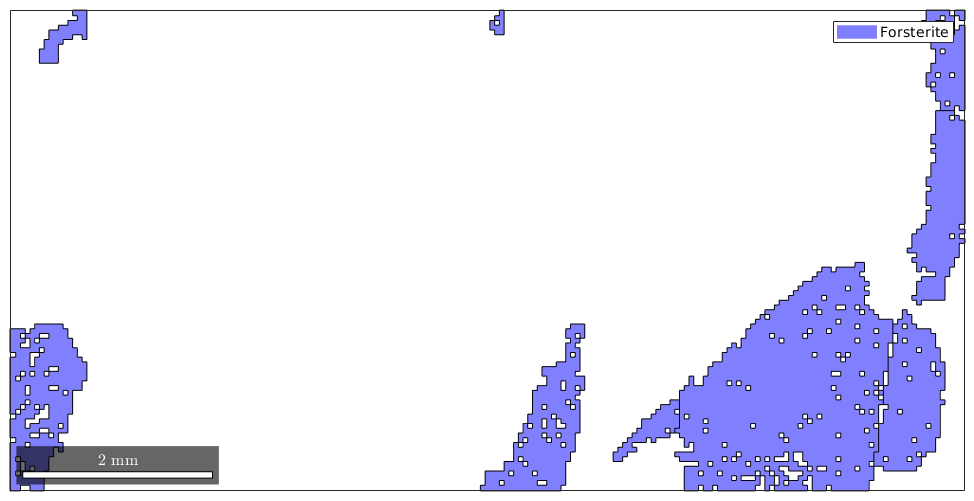
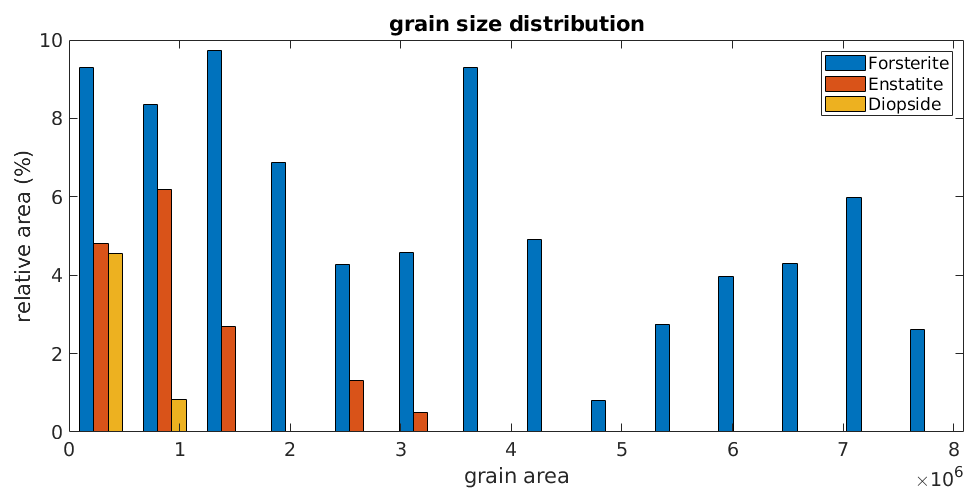
Then the following command gives you a nice overview over the grain size distributions of the grains
hist(grains)
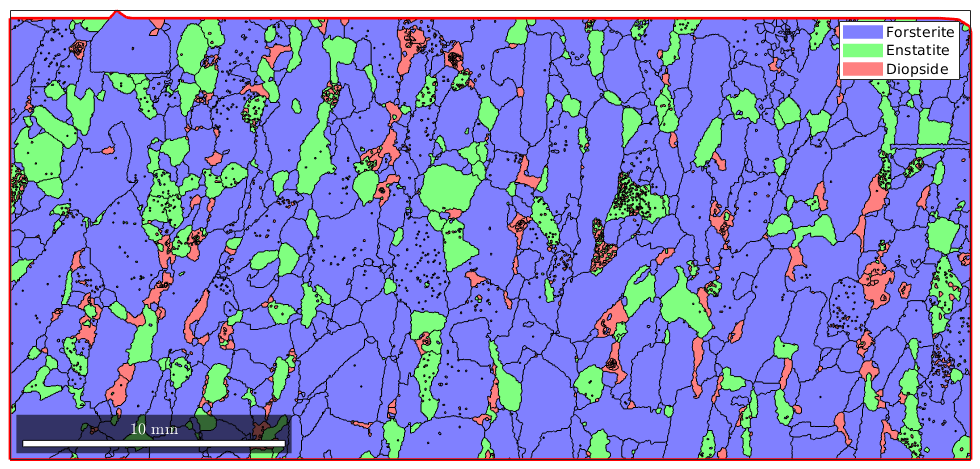
Sometimes it is desirable to remove all boundary grains as they might distort grain statistics. To do so one should remember that each grain boundary has a property grainId which stores the ids of the neigbouring grains. In the case of an outer grain boundary, one of the neighbouring grains has the id zero. We can filter out all these boundary segments by
% ids of the outer boundary segment outerBoundary_id = any(grains.boundary.grainId==0,2); plot(grains) hold on plot(grains.boundary(outerBoundary_id),'linecolor','red','linewidth',2) hold off
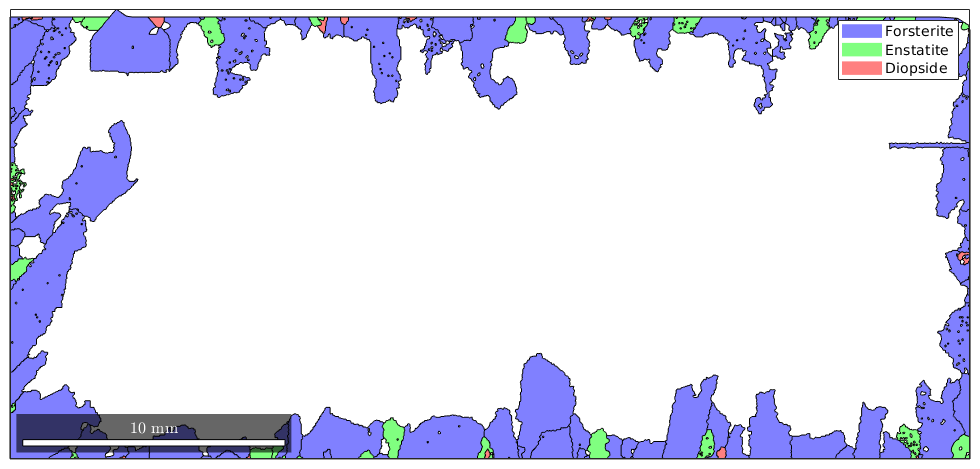
Now grains.boundary(outerBoundary_id).grainId is a list of grain ids where the first column is zero, indicating the outer boundary, and the second column contains the id of the boundary grain. Hence, it remains to remove all grains with these ids.
% next we compute the corresponding grain_id grain_id = grains.boundary(outerBoundary_id).grainId; % remove all zeros grain_id(grain_id==0) = []; % and plot the boundary grains plot(grains(grain_id))
finally, we can remove the boundary grains by
grains(grain_id) = [] plot(grains)
grains = grain2d
Phase Grains Pixels Mineral Symmetry Crystal reference frame
1 968 116882 Forsterite mmm
2 480 23877 Enstatite mmm
3 1467 8871 Diopside 12/m1 X||a*, Y||b*, Z||c
boundary segments: 40105
triple points: 3383
Properties: GOS, meanRotation
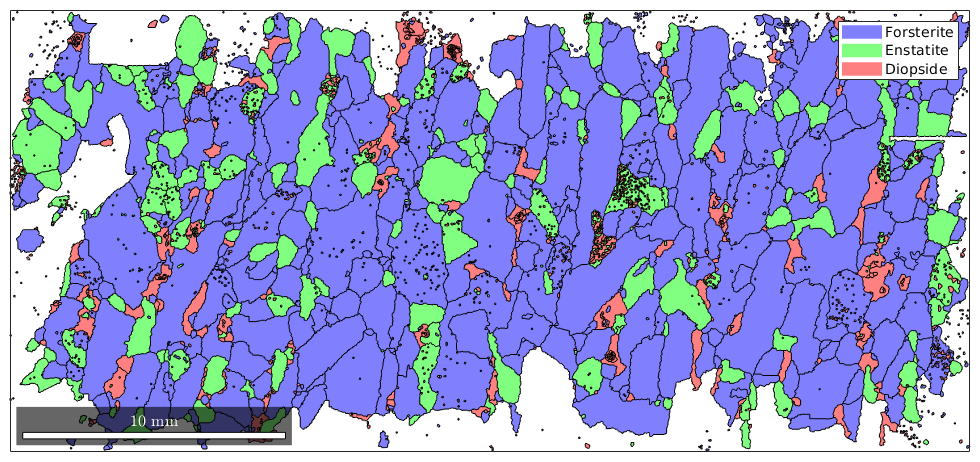
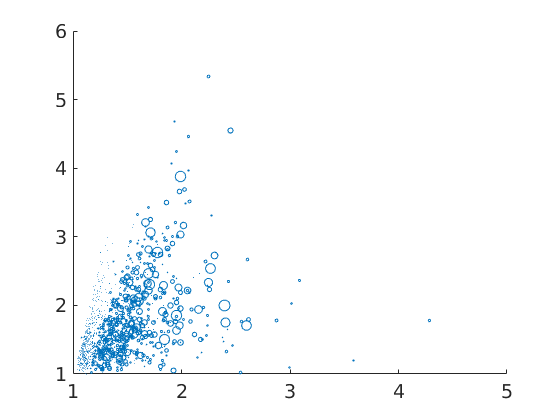
Beside the area, there are various other geometric properties that can be computed for grains, e.g., the perimeter, the diameter, the equivalentRadius, the equivalentPerimeter, the aspectRatio, and the shapeFactor. The following is a simple scatter plot of shape factor against aspect ratio to check for correlation.
% the size of the dots corresponds to the area of the grains close all scatter(grains.shapeFactor, grains.aspectRatio, 70*grains.area./max(grains.area))
plot(grains,log(grains.aspectRatio))
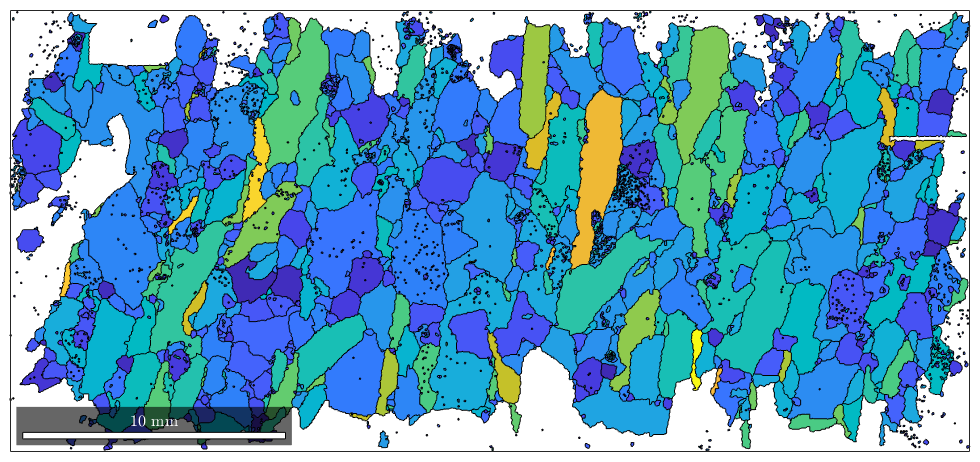
Spatial Dependencies
One interesting question would be, whether a polyphase system has dependence in the spatial arrangement or not, therefore, we can count the transitions to a neighbour grain
%[J, T, p ] = joinCount(grains,grains.phase)
| DocHelp 0.1 beta |