Plotting spatially indexed EBSD data
How to visualize EBSD data
This section gives you an overview of the functionality MTEX offers to visualize spatial orientation data.
| On this page ... |
| Phase Plots |
| Visualizing arbitrary properties |
| Visualizing orientations |
| Customizing the color |
| Coloring certain fibres |
| Coloring certain orientations |
| Combining different plots |
Phase Plots
Let us first import some EBSD data with a script file
close all; plotx2east mtexdata forsterite csFo = ebsd('Forsterite').CS
csFo = crystalSymmetry mineral : Forsterite color : light blue symmetry: mmm a, b, c : 4.8, 10, 6
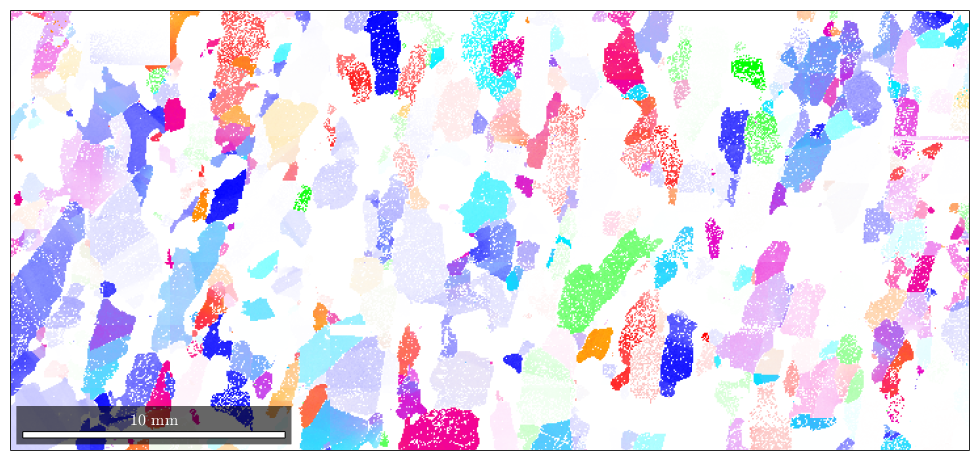
By default, MTEX plots a phase map for EBSD data.
plot(ebsd)
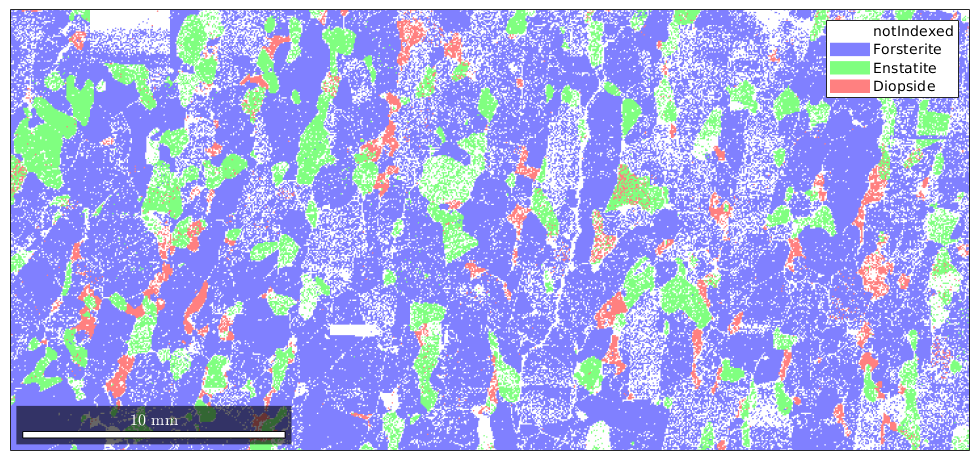
You can access the color of each phase by
ebsd('Diopside').colorans =
1.0000 0.5000 0.5000
These values are RGB values, e.g. to make the color for diopside even more red we can do
ebsd('Diopside').color = [1 0 0]; plot(ebsd('indexed'))
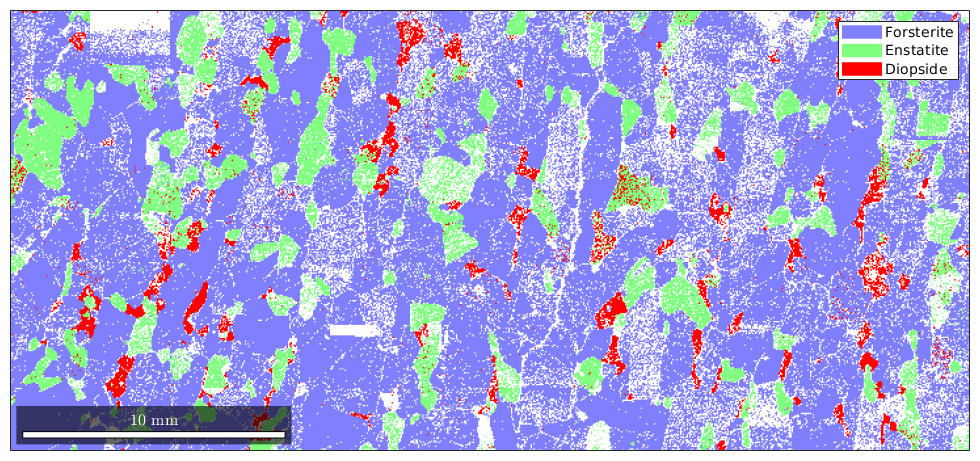
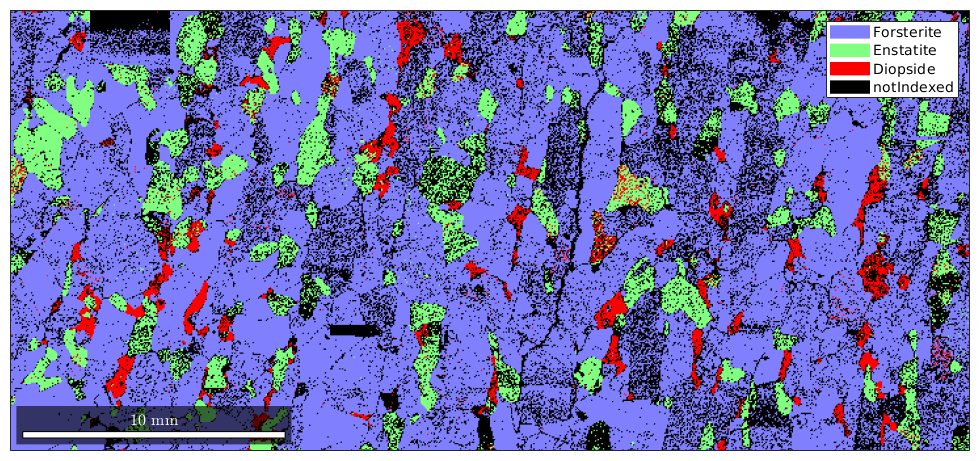
By default, not indexed phases are plotted as white. To directly specify a color for some EBSD data use the option FaceColor.
hold on plot(ebsd('notIndexed'),'FaceColor','black') hold off
Visualizing arbitrary properties
Apart from the phase information, we can use any other property to colorize the EBSD data. As an example, we may plot the band contrast
plot(ebsd,ebsd.bc) colormap gray % this makes the image grayscale mtexColorbar
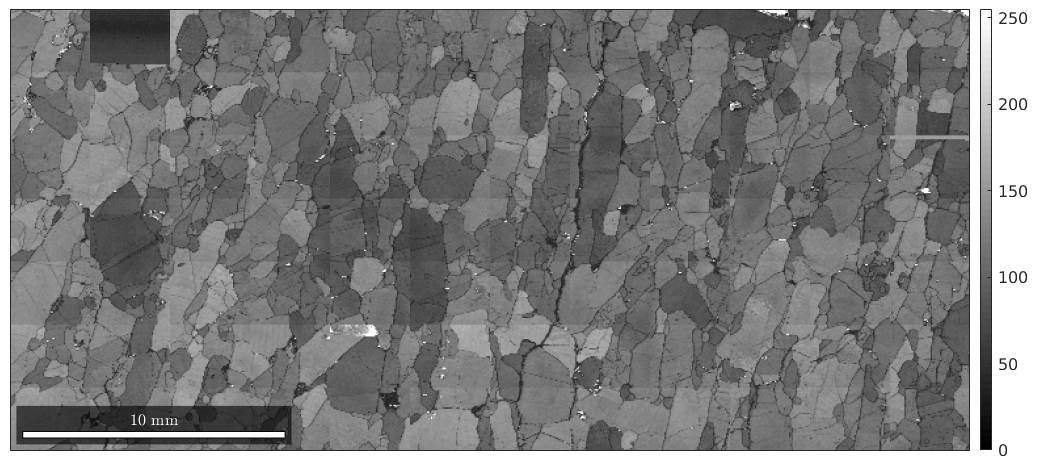
Visualizing orientations
Actually, we can pass any list of numbers or colors as a second input argument to be visualized together with the ebsd data. In order to visualize orientations in an EBSD map, we have first to compute a color for each orientation. The most simple way is to assign to each orientation its rotational angle. This is done by the command
plot(ebsd('Forsterite'),ebsd('Forsterite').orientations.angle./degree) mtexColorbar
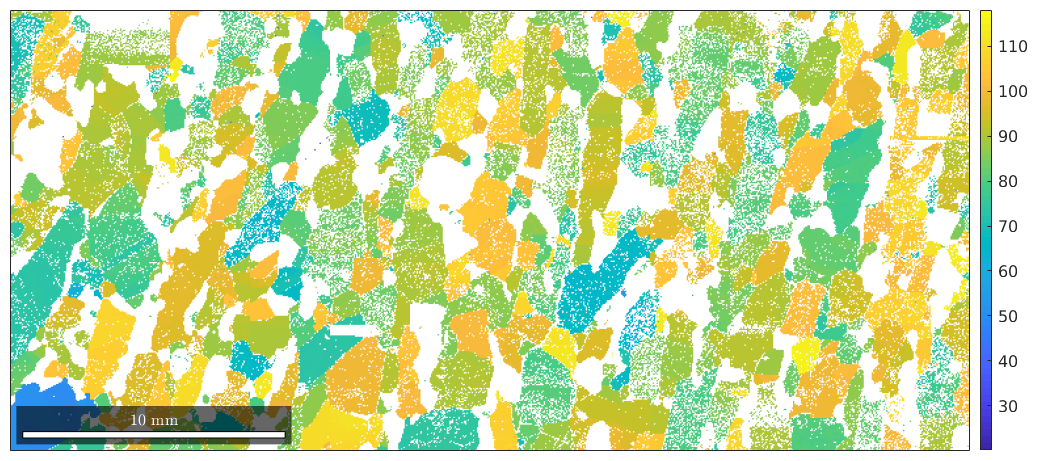
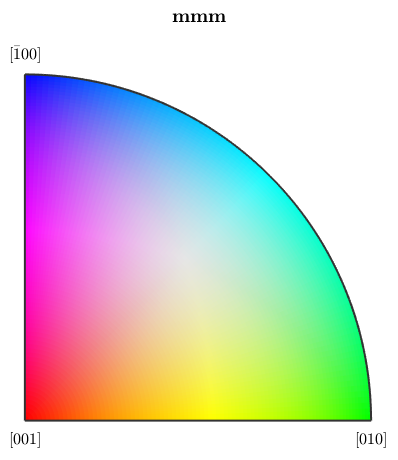
Obviously, the rotational angle is not a very distintive representative for orientations. A more common approach is to define a colorization of the fundamental secor of the inverse pole figure, a so called ipf color key, and to colorize orientations according to their position in a fixed inverse pole figure. Let's consider the following standard key.
% this defines an ipf color key for the Forsterite phase ipfKey = ipfHSVKey(ebsd('Forsterite')) % this is the colored fundamental sector plot(ipfKey)
ipfKey =
ipfHSVKey with properties:
colorPostRotation: [1×1 rotation]
colorStretching: 1
whiteCenter: [1×1 vector3d]
grayValue: [0.2000 0.5000]
grayGradient: 0.5000
maxAngle: Inf
inversePoleFigureDirection: [1×1 vector3d]
dirMap: [1×1 HSVDirectionKey]
CS1: [4×2 crystalSymmetry]
CS2: [1×1 specimenSymmetry]
antipodal: 0
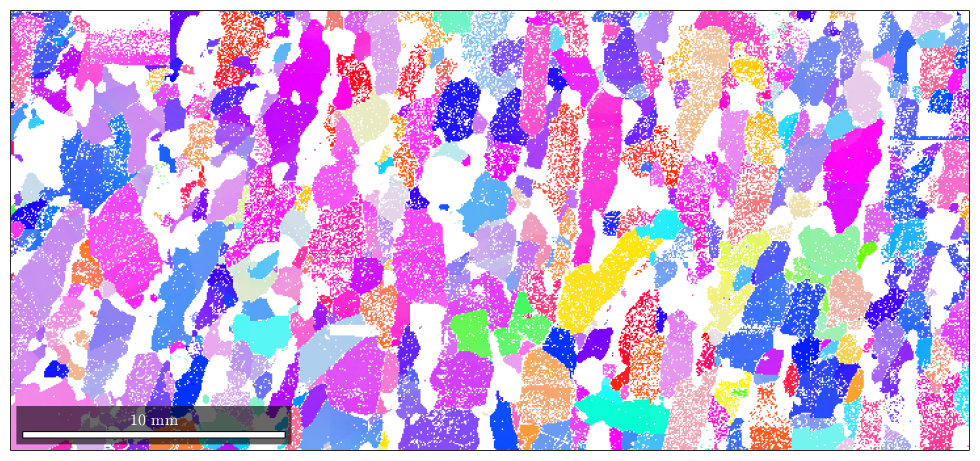
In order to apply this color key for colorizing the map we proceed as follows
% compute the colors color = ipfKey.orientation2color(ebsd('Fo').orientations); % plot the colors close all plot(ebsd('Forsterite'),color)
Orientation color key usually provide several options to alter the alignment of colors. Let's give some examples
% we may interchange green and blue by setting
ipfKey.colorPostRotation = reflection(yvector);
plot(ipfKey)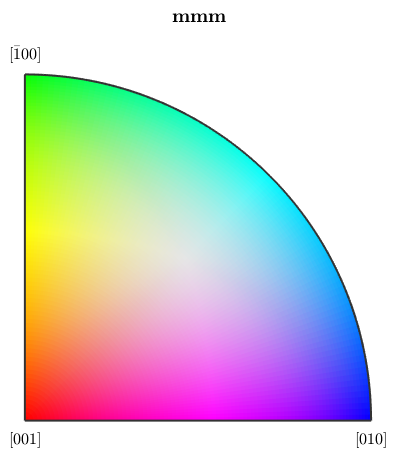
or cycle of colors red, green, blue by
ipfKey.colorPostRotation = rotation.byAxisAngle(zvector,120*degree); plot(ipfKey)
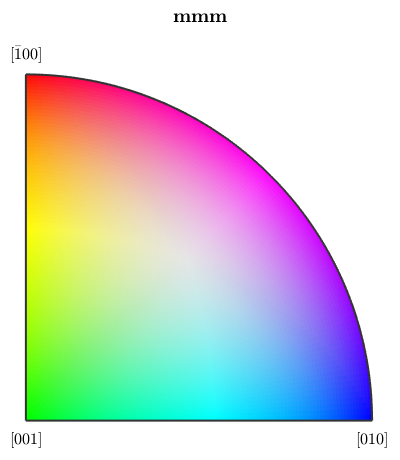
Furthermore, we can explicitly set the inverse pole figure directions by
ipfKey.inversePoleFigureDirection = zvector; % compute the colors again color = ipfKey.orientation2color(ebsd('Forsterite').orientations); % and plot them close all plot(ebsd('Forsterite'),color)
Besides the recommended color key ipfHSVKey MTEX includes a bunch of other color keys
Customizing the color
In some cases, it might be useful to color certain orientations after one needs. This can be done in two ways, either to color a certain fibre or a certain orientation.
Coloring certain fibres
To color a fibre, one has to specify the crystal direction h together with its RGB color and the specimen direction r, which should be marked.
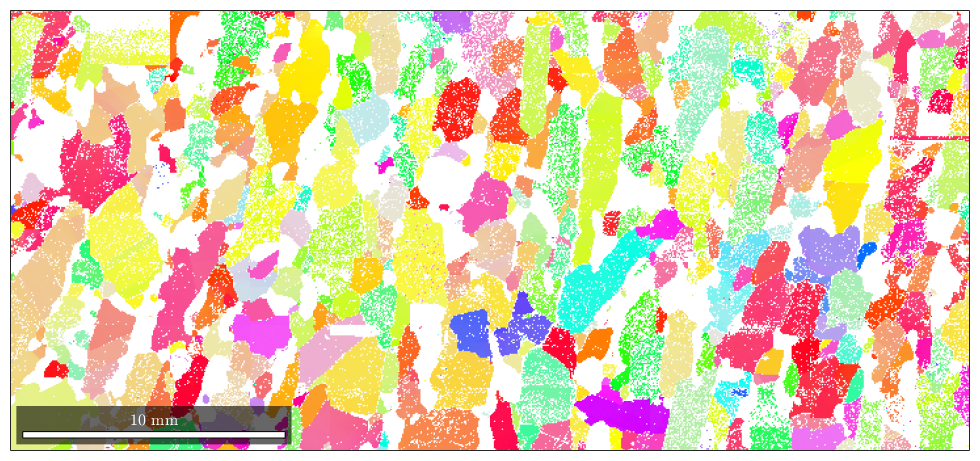
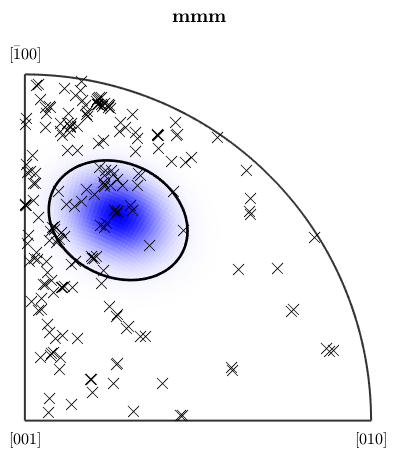
% define a fibre f = fibre(Miller(1,1,1,csFo),zvector); % set up coloring ipfKey = ipfSpotKey(csFo); ipfKey.inversePoleFigureDirection = f.r; ipfKey.center = f.h; ipfKey.color = [0 0 1]; ipfKey.psi = deLaValleePoussinKernel('halfwidth',7.5*degree); plot(ebsd('fo'),ipfKey.orientation2color(ebsd('fo').orientations))
the option halfwidth controls half of the intensity of the color at a given distance. Here we have chosen the (111)[001] fibre to be drawn in blue, and at 7.5 degrees, where the blue should be only lighter.
plot(ipfKey) hold on circle(f.h.project2FundamentalRegion,15*degree,'linewidth',2)
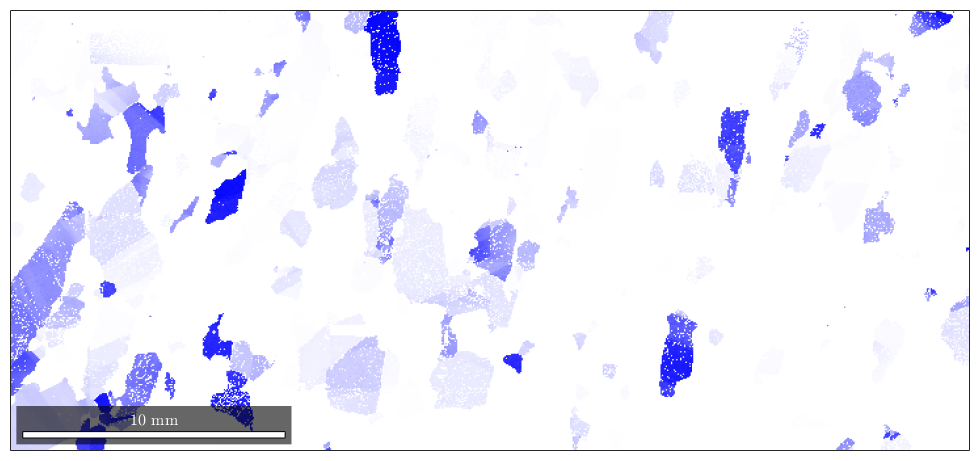
the percentage of blue colored area in the map is equivalent to the fibre volume
vol = volume(ebsd('fo').orientations,f,15*degree) plotIPDF(ebsd('fo').orientations,zvector,'markercolor','k','marker','x','points',200) hold off
vol =
0.2480
I'm plotting 200 random orientations out of 152345 given orientations
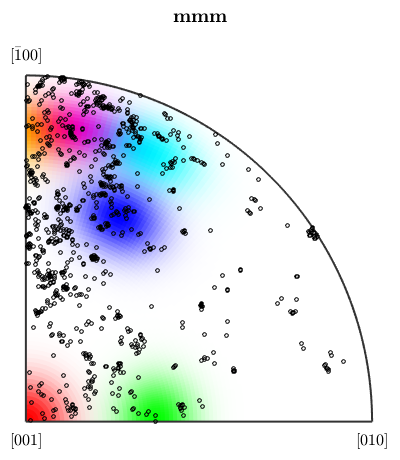
we can easily extend the colorcoding
% the centers in the inverse pole figure ipfKey.center = Miller({0 0 1},{0 1 1},{1 1 1},{11 4 4},{5 0 2},{5 5 2},csFo); % the correspnding collors ipfKey.color = [[1 0 0];[0 1 0];[0 0 1];[1 0 1];[1 1 0];[0 1 1]]; % plot the key plot(ipfKey) hold on plot(ebsd('fo').orientations,'MarkerFaceColor','none','MarkerEdgeColor','k','MarkerSize',3,'points',1000) hold off
I'm plotting 1000 random orientations out of 152345 given orientations
close all; plot(ebsd('fo'),ipfKey.orientation2color(ebsd('fo').orientations))
Coloring certain orientations
We might be interested in locating some special orientation in our orientation map. The definition of colors for certain orientations is carried out similarly as in the case of fibres
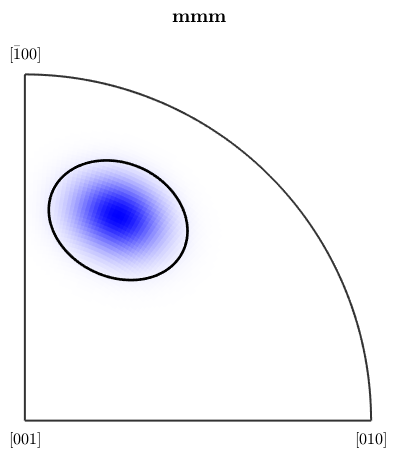
ipfKey = spotColorKey(ebsd('Fo')); ipfKey.center = mean(ebsd('Forsterite').orientations,'robust'); ipfKey.color = [0,0,1]; ipfKey.psi = deLaValleePoussinKernel('halfwidth',20*degree); plot(ebsd('fo'),ipfKey.orientation2color(ebsd('fo').orientations)) % and the correspoding colormap figure(2) plot(ipfKey,'sections',9,'sigma')
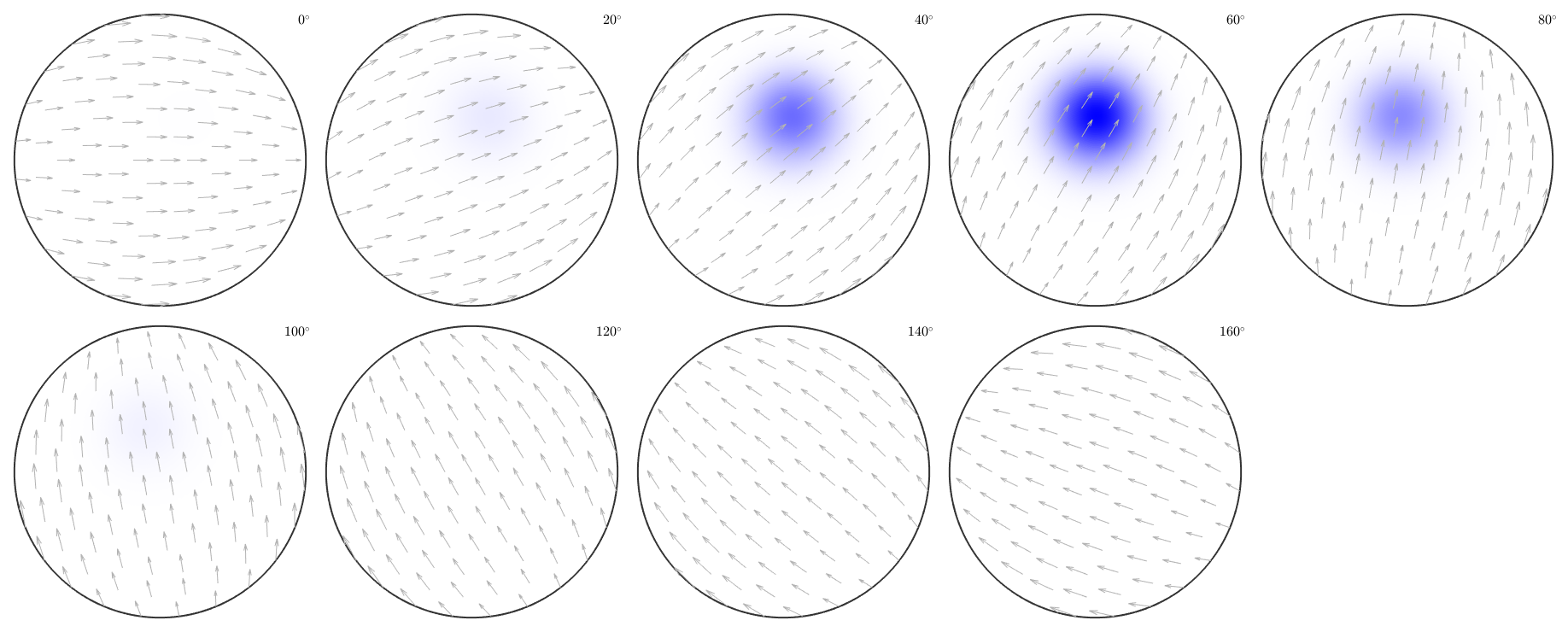
the area of the colored EBSD data in the map corresponds to the volume portion (in percent)
vol = 100 * volume(ebsd('fo').orientations,ipfKey.center,20*degree)vol =
1.3227
actually, the colored measurements stress a peak in the ODF
close all odf = calcODF(ebsd('fo').orientations,'halfwidth',10*degree,'silent'); plot(odf,'sections',9,'silent','sigma') mtexColorbar
Warning: Plot properties not compatible to previous plot! I'going to create a new figure.
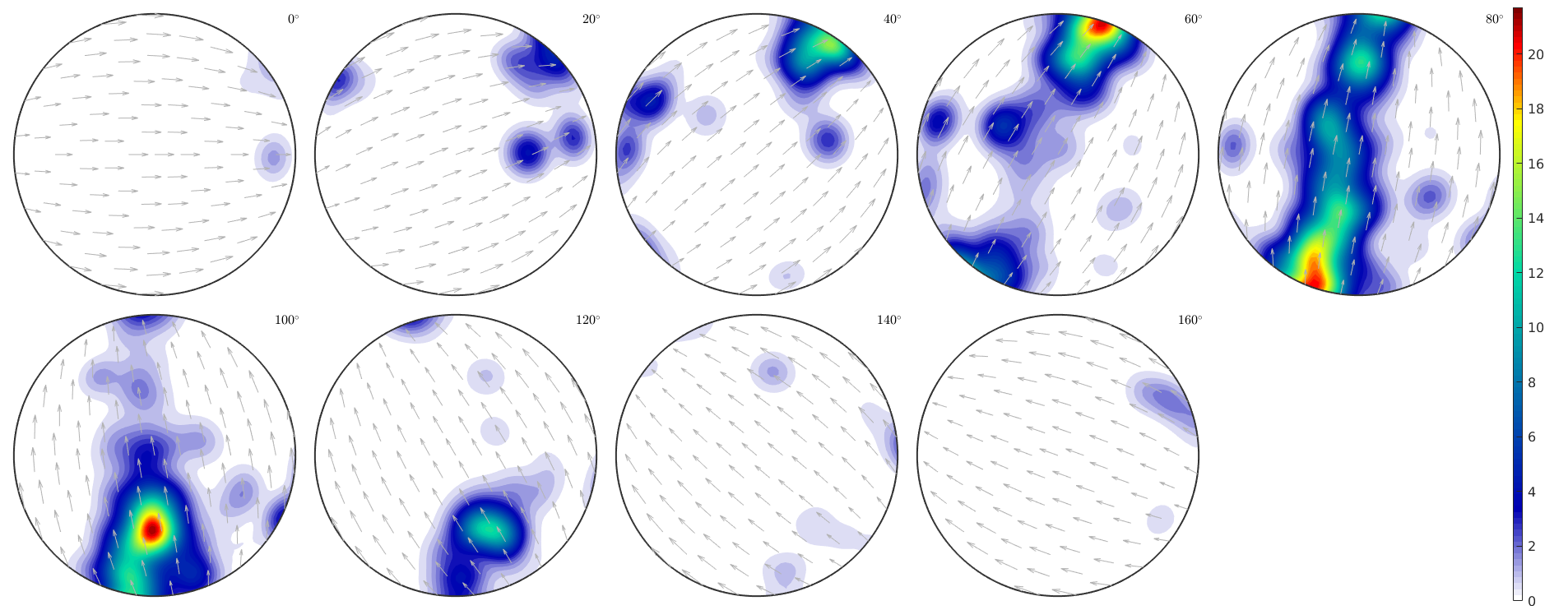
Combining different plots
Combining different plots can be done either by plotting only subsets of the EBSD data or via the option 'faceAlpha'. Note that the option 'faceAlpha' requires the renderer of the figure to be set to 'opengl'.
close all; plot(ebsd,ebsd.bc,'micronbar','off') mtexColorMap black2white ipfKey = ipfSpotKey(csFo); ipfKey.inversePoleFigureDirection = zvector; ipfKey.center = Miller(1,1,1,csFo); ipfKey.color = [0 0 1]; ipfKey.psi = deLaValleePoussinKernel('halfwidth',7.5*degree); hold on plot(ebsd('fo'),ipfKey.orientation2color(ebsd('fo').orientations),'FaceAlpha',0.5) hold off
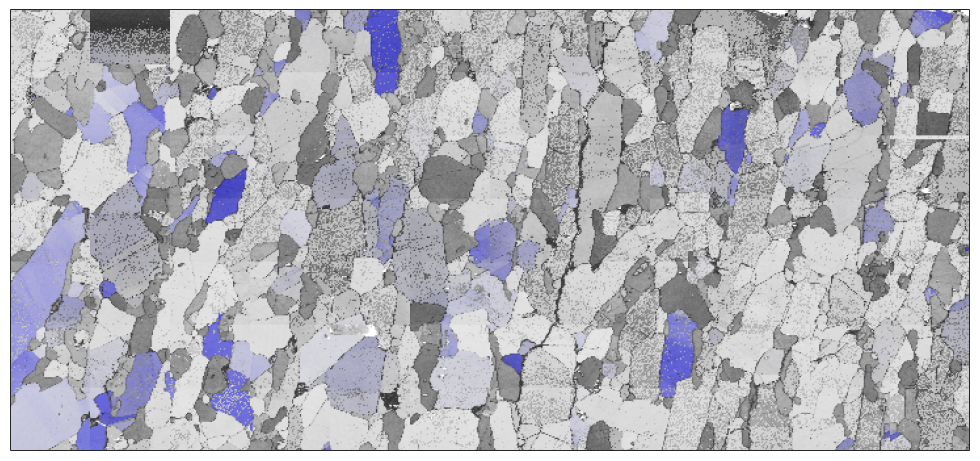
another example
close all; plot(ebsd,ebsd.bc) mtexColorMap black2white hold on plot(ebsd('fo'),'FaceAlpha',0.5) hold off
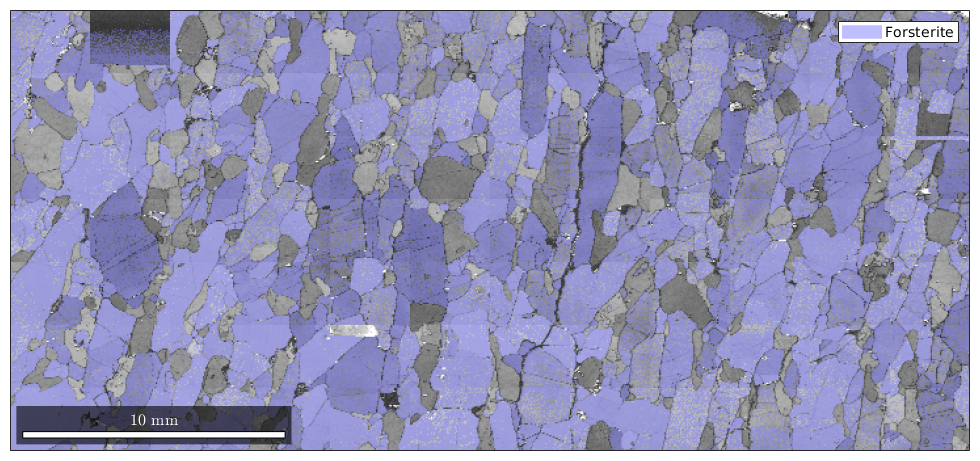
| DocHelp 0.1 beta |